Anti-Tumor Activity of Novel Nimotuzumab-Functionalized Gold Nanoparticles as a Potential Immunotherapeutic Agent against Skin and Lung Cancers
Abstract
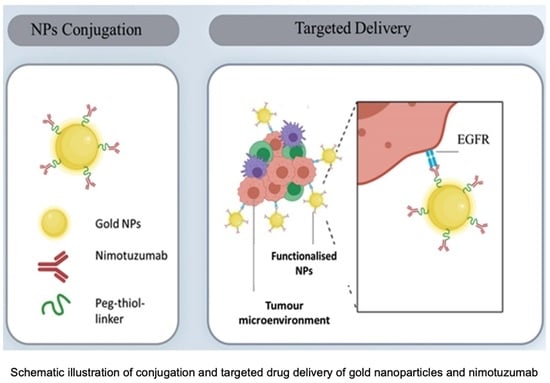
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Synthesis of Spherical Citrate–Tannate-Capped AuNPs
2.2.2. PEGylation of NmAb and AuNPs and Surface Functionalization of AuNPs (AuNP-NmAb)
2.2.3. Characterization of Nanoparticles
Transmission Electron Microscope (TEM)
Dynamic Light Scattering (DLS)
Nanoparticle Tracking Analysis (NTA)
Fourier-Transform Infrared Spectrometer (FTIR)
Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
UV-Vis Spectroscopy
Colloidal Stability in Salt and a Biological Medium by UV-Vis and DLS
Calculation of Antibody Aggregation and Antibody-Binding Percentage through UV-Vis Spectroscopy
2.2.4. In Vitro Studies
Culture of A431 and A549 Cells
Determination of the Percentage of Cell Viability through MTT Assay
Estimation of Cellular Uptake of AuNPs through ICP-MS Analysis
Statistical Analysis
3. Results
3.1. AuNP Synthesis and Surface Functionalization with NmAb
3.2. Physicochemical Characterization of PEGylated Bare AuNPs and NmAb-Functionalized AuNPs (AuNP-NmAb)
3.3. Determination of the Surface Functionalization of AuNPs with NmAb and Their Stability Analysis
3.4. In Vitro Studies
3.4.1. Evaluation of Anti-Tumor Activity in EGFR+ Cancer Cells
3.4.2. Evaluation of the Cellular Uptake of AuNP-NmAb in EGFR+ Cancer Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, F.; Wang, S.; Yin, L.; Yang, Y.; Guan, Y.; Wang, W.; Xu, H.; Tao, N. Quantification of epidermal growth factor receptor expression level and binding kinetics on cell surfaces by surface plasmon resonance imaging. Anal. Chem. 2015, 87, 9960–9965. [Google Scholar] [CrossRef] [PubMed]
- Wee, P.; Wang, Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Gandhi, S.; Tokumaru, Y.; Yan, L.; Yamada, A.; Matsuyama, R.; Ishikawa, T.; Endo, I.; Takabe, K. Conflicting roles of EGFR expression by subtypes in breast cancer. Am. J. Cancer Res. 2021, 11, 5094–5110. [Google Scholar] [PubMed]
- Nair, S.; Bonner, J.A.; Bredel, M. EGFR mutations in head and neck squamous cell carcinoma. Int. J. Mol. Sci. 2022, 23, 3818. [Google Scholar] [CrossRef]
- London, M.; Gallo, E. Epidermal growth factor receptor (EGFR) involvement in epithelial-derived cancers and its current antibody-based immunotherapies. Cell Biol. Int. 2020, 44, 1267–1282. [Google Scholar] [CrossRef]
- Mazorra, Z.; Chao, L.; Lavastida, A.; Sanchez, B.; Ramos, M.; Iznaga, N.; Crombet, T. Nimotuzumab: Beyond the EGFR signaling cascade inhibition. Semin. Oncol. 2018, 45, 18–26. [Google Scholar] [CrossRef]
- Cai, W.Q.; Zeng, L.S.; Wang, L.F.; Wang, Y.Y.; Cheng, J.T.; Zhang, Y.; Han, Z.W.; Zhou, Y.; Huang, S.L.; Wang, X.W.; et al. The latest battles between EGFR monoclonal antibodies and resistant tumor cells. Front. Oncol. 2020, 10, 1249. [Google Scholar] [CrossRef]
- Mazorra, Z.; Lavastida, A.; Concha-Benavente, F.; Valdés, A.; Srivastava, R.M.; García-Bates, T.M.; Hechavarría, E.; González, Z.; González, A.; Lugiollo, M.; et al. Nimotuzumab induces NK cell activation, cytotoxicity, dendritic cell maturation and expansion of EGFR-specific t cells in head and neck cancer patients. Front. Pharmacol. 2017, 8, 382. [Google Scholar] [CrossRef]
- Fauvel, B.; Yasri, A. Antibodies directed against receptor tyrosine kinases: Current and future strategies to fight cancer. MAbs 2014, 6, 838–851. [Google Scholar] [CrossRef]
- Sidaway, P. EGFR inhibition is effective against KRAS-wild-type disease. Nat. Rev. Clin. Oncol. 2017, 14, 525. [Google Scholar] [CrossRef]
- Lee, C.M.; Tannock, I.F. The distribution of the therapeutic monoclonal antibodies cetuximab and trastuzumab within solid tumors. BMC Cancer 2010, 10, 255. [Google Scholar] [CrossRef] [PubMed]
- Cruz, E.; Veysel, K. Synthesis and enhanced cellular uptake in vitro of anti-HER2 multifunctional gold nanoparticle. Cancers 2019, 11, 870. [Google Scholar] [CrossRef] [PubMed]
- Viswanadh, M.K.; Vikas; Jha, A.; Adena, S.K.R.; Mehata, A.K.; Priya, V.; Neogi, K.; Poddar, S.; Mahto, S.K.; Muthu, M.S. Formulation and in vivo efficacy study of cetuximab decorated targeted bioadhesive nanomedicine for non-small-cell lung cancer therapy. Nanomedicine 2020, 15, 2345–2367. [Google Scholar] [CrossRef] [PubMed]
- Julien, D.C.; Behnke, S.; Wang, G.; Murdoch, G.K.; Hill, R.A. Utilization of monoclonal antibody-targeted nanomaterials in the treatment of cancer. MAbs 2011, 3, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Steckiewicz, K.P.; Barcinska, E.; Malankowska, A.; Zauszkiewicz-Pawlak, A.; Nowaczyk, G.; Zaleska-Medynska, A.; Inkielewicz-Stepniak, I. Impact of gold nanoparticles shape on their cytotoxicity against human osteoblast and osteosarcoma in in vitro model. Evaluation of the safety of use and anti-cancer potential. J. Mater. Sci. Mater. Med. 2019, 30, 22. [Google Scholar] [CrossRef]
- Sengupta, A.; Azharuddin, M.; Al-Otaibi, N.; Hinkula, J. Efficacy and immune response elicited by gold nanoparticle- based nanovaccines against infectious diseases. Vaccines 2022, 10, 505. [Google Scholar] [CrossRef]
- Wang, Y.; Quinsaat, J.E.Q.; Ono, T.; Maeki, M.; Tokeshi, M.; Isono, T.; Tajima, K.; Satoh, T.; Sato, S.-I.; Miura, Y.; et al. Enhanced dispersion stability of gold nanoparticles by the physisorption of cyclic poly(ethylene glycol). Nat. Commun. 2020, 11, 6089. [Google Scholar] [CrossRef]
- Eck, W.; Craig, G.; Sigdel, A.; Ritter, G.; Old, L.J.; Tang, L.; Brennan, M.F.; Allen, P.J.; Mason, M.D. PEGylated gold nanoparticle conjugated to monoclonal f19 antibodies as targeted labeling agents for human pancreatic carcinoma tissue. ACS Nano 2008, 2, 2263–2272. [Google Scholar] [CrossRef]
- Goddard, Z.R.; Marín, M.J.; Russell, D.A.; Searcey, M. Active targeting of gold nanoparticle as cancer therapeutics. Chem. Soc. Rev. 2020, 49, 8774–8789. [Google Scholar] [CrossRef]
- Chanda, N.; Kattumuri, V.; Shukla, R.; Zambre, A.; Katti, K.; Upendran, A.; Kulkarni, R.R.; Kan, P.; Fent, G.M.; Casteel, S.W.; et al. Bombesin functionalised gold nanoparticle show in vitro and in vivo cancer receptor specificity. Proc. Natl. Acad. Sci. USA 2010, 107, 8760–8765. [Google Scholar] [CrossRef]
- Niikura, K.; Matsunaga, T.; Suzuki, T.; Kobayashi, S.; Yamaguchi, H.; Orba, Y.; Kawaguchi, A.; Hasegawa, H.; Kajino, K.; Ninomiya, T.; et al. Gold Nanoparticle as a Vaccine Platform: Influence of Size and Shape on Immunological Responses in Vitro and in Vivo. ACS Nano 2013, 7, 3926–3938. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Qoutah, W.W.; Free, P.; Hobley, J.; Fernig, D.G.; Paramelle, D. Features of thiolated ligands promoting resistance to ligand exchange in self-assembled monolayers on gold nanoparticle. Aust. J. Chem. 2012, 65, 266. [Google Scholar] [CrossRef]
- Yeo, E.L.L.; Chua, A.J.S.; Parthasarathy, K.; Yeo, H.Y.; Ng, M.L.; Kah, J.C.Y. Understanding aggregation-based assays: Nature of protein corona and number of epitopes on antigen matters. RSC Adv. 2015, 5, 14982–14993. [Google Scholar] [CrossRef]
- Freitas, S.; Martins, R.; Campos, A.; Azevedo, J.; Osório, H.; Costa, M.; Barros, P.; Vasconcelos, V.; Urbatzka, R. Insights into the potential of picoplanktonic marine cyanobacteria strains for cancer therapies—Cytotoxic mechanisms against the RKO colon cancer cell line. Toxicon 2016, 119, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Oltolina, F.; Colangelo, D.; Miletto, I.; Clemente, N.; Miola, M.; Verné, E.; Prat, M.; Follenzi, A. Tumor targeting by monoclonal antibody functionalised magnetic nanoparticle. Nanomaterials 2019, 9, 1575. [Google Scholar] [CrossRef]
- Borse, V.; Konwar, A.N. Synthesis, and characterization of gold nanoparticle as a sensing tool for the lateral flow immunoassay development. Sens. Int. 2020, 1, 100051. [Google Scholar] [CrossRef]
- Aromal, S.A.; Philip, D. Facile one-pot synthesis of gold nanoparticle using tannic acid and its application in catalysis. Phys. E Low-Dimens. Syst. Nanostruct. 2012, 44, 1692–1696. [Google Scholar] [CrossRef]
- Ranoszek-Soliwoda, K.; Tomaszewska, E.; Socha, E.; Krzyczmonik, P.; Ignaczak, A.; Orlowski, P.; Krzyzowska, M.; Celichowski, G.; Grobelny, J. The role of tannic acid and sodium citrate in the synthesis of silver nanoparticle. J. Nanopart. Res. 2017, 19, 273. [Google Scholar] [CrossRef]
- Hermanson, G. PEGylation, and synthetic polymer modification. In Bioconjugate Techniques; Elsevier: Amsterdam, The Netherlands, 2013; pp. 787–838. [Google Scholar] [CrossRef]
- Mosquera, J.M.; Henriksen-Lacey, M.; García, I.; Martínez-Calvo, M.; Rodríguez, J.; Mascareñas, J.L.; Liz-Marzán, L.M. Cellular uptake of gold nanoparticle triggered by host-guest interactions. J. Am. Chem. Soc. 2018, 140, 4469–4472. [Google Scholar] [CrossRef]
- Wu, J.; Zheng, Y.; Liu, M.; Shan, W.; Zhang, Z.; Huang, Y. Biomimetic virus-like and charge reversible nanoparticle to sequentially overcome mucus and epithelial barriers for oral insulin delivery. ACS Appl. Mater. Interfaces 2018, 10, 9916–9928. [Google Scholar] [CrossRef]
- Gustafson, H.H.; Holt-Casper, D.; Grainger, D.W.; Ghandehari, H. Nanoparticle uptake: The phagocyte problem. Nano Today 2015, 10, 487–510. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.; Kantner, K.; Zhang, Q.; Soliman, M.G.; Del Pino, P.; Parak, W.J.; Onur, M.A.; Valdeperez, D.; Rejman, J.; Pelaz, B. Conjugation of polymer-coated gold nanoparticle with antibodies—Synthesis and characterization. Nanomaterials 2015, 5, 1297–1316. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, R.B.; Ragupathi, A.; Dyer, R.B. Characterizing the Surface Coverage of Protein–Gold Nanoparticle Bioconjugates. Bioconjug. Chem. 2018, 29, 2691–2700. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cai, R.; Kawazoe, N.; Chen, G. Facile preparation of albumin-stabilized gold nanostars for the targeted photothermal ablation of cancer cells. J. Mater. Chem. B 2015, 3, 5806–5814. [Google Scholar] [CrossRef]
- Suk, J.S.; Xu, Q.; Kim, N.; Hanes, J.; Ensign, L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016, 99, 28–51. [Google Scholar] [CrossRef]
- Oh, E.; Susumu, K.; Goswami, R.; Mattoussi, H. One-phase synthesis of water-soluble gold nanoparticle with control over the size and surface functionalities. Langmuir 2010, 26, 7604–7613. [Google Scholar] [CrossRef]
- Ohnishi, S.; Murata, M.; Hato, M. Correlation between surface morphology and surface forces of protein A adsorbed on mica. Biophys. J. 1998, 74, 455–465. [Google Scholar] [CrossRef]
- Jans, H.; Liu, X.; Austin, L.; Maes, G.; Huo, Q. Dynamic light scattering as a powerful tool for gold nanoparticle bioconjugation and biomolecular binding studies. Anal. Chem. 2009, 81, 9425–9432. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.K.; Aveyard, J.; Fernig, D.G. Determination of size and concentration of gold nanoparticle from UV−Vis spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef]
- Aldewachi, H.; Woodroofe, N.; Gardiner, P. Study of the stability of functionalised gold nanoparticle for the colorimetric detection of dipeptidyl peptidase iv. Appl. Sci. 2018, 8, 2589. [Google Scholar] [CrossRef]
- Puertas, S.; Moros, M.; Fernández-Pacheco, R.; Ibarra, M.R.; Grazú, V.; de-la-Fuente, J.M. Designing novel nano-immunoassays: Antibody orientation versus sensitivity. J. Phys. D Appl. Phys. 2010, 43, 474012. [Google Scholar] [CrossRef]
- Clarizia, L.-J.A.; Sok, D.; Wei, M.; Mead, J.; Barry, C.; McDonald, M.J. Antibody orientation enhanced by selective polymer–protein noncovalent interactions. Anal. Bioanal. Chem. 2008, 393, 1531. [Google Scholar] [CrossRef] [PubMed]
- Boland, W.K.; Bebb, G. Nimotuzumab: A novel anti-EGFR monoclonal antibody that retains anti-EGFR activity while minimizing skin toxicity. Expert Opin. Biol. Ther. 2009, 9, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Garrido, G.; Tikhomirov, I.A.; Rabasa, A.; Yang, E.; Gracia, E.; Iznaga, N.; Fernández, L.E.; Crombet, T.; Kerbel, R.S.; Pérez, R. Bivalent binding by the intermediate affinity of nimotuzumab: A contribution to explain antibody clinical profile. Cancer Biol. Ther. 2011, 11, 373–382. [Google Scholar] [CrossRef]
- Sylvester, P.W. Optimization of the tetrazolium dye (MTT) colorimetric assay for cellular growth and viability. In Drug Design and Discovery: Methods and Protocols; Satyanarayanajois, S.D., Ed.; Humana Press: Totowa, NJ, USA, 2011; pp. 157–168. [Google Scholar] [CrossRef]
- Suman, S.; Priya, R.; Kameswaran, M. Induction of different cellular arrest and molecular responses in low EGFR expressing A549 and high EGFR expressing A431 tumor cells treated with various doses of 177Lu-Nimotuzumab. Int. J. Radiat. Biol. 2020, 96, 1144–1156. [Google Scholar] [CrossRef]
- Iram, S.; Zahera, M.; Wahid, I.; Abu Baker, A.; Raish, M.; Khan, A.; Ali, N.; Ahmad, S.; Khan, M.S. Cisplatin bioconjugated enzymatic GNPs amplify the effect of cisplatin with acquiescence. Sci. Rep. 2019, 9, 13826. [Google Scholar] [CrossRef]
- Manzano, C.M.; Nakahata, D.H.; de Paiva, R.E.F. Revisiting metallodrugs for the treatment of skin cancers. Coord. Chem. Rev. 2022, 462, 214506. [Google Scholar] [CrossRef]
- Agnello, L.; Tortorella, S.; d’Argenio, A.; Carbone, C.; Camorani, S.; Locatelli, E.; Auletta, L.; Sorrentino, D.; Fedele, M.; Zannetti, A.; et al. Optimizing cisplatin delivery to triple-negative breast cancer through novel EGFR aptamer-conjugated polymeric nanovectors. J. Exp. Clin. Cancer Res. 2021, 40, 239. [Google Scholar] [CrossRef] [PubMed]
- Alkilany, A.; Murphy, C. Toxicity, and cellular uptake of gold nanoparticle: What we have learned so far? J. Nanopart. Res. 2010, 12, 2313–2333. [Google Scholar] [CrossRef]
- Kao, H.-W.; Lin, Y.-Y.; Chen, C.-C.; Chi, K.-H.; Tien, D.-C.; Hsia, C.-C.; Lin, W.-J.; Chen, F.-D.; Lin, M.-H.; Wang, H.-E. Biological characterization of cetuximab-conjugated gold nanoparticle in a tumor animal model. Nanotechnology 2014, 25, 295102. [Google Scholar] [CrossRef] [PubMed]
- El Hallal, R.; Lyu, N.; Wang, Y. Effect of cetuximab-conjugated gold nanoparticle on the cytotoxicity and phenotypic evolution of colorectal cancer cells. Molecules 2021, 26, 567. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Jiang, C.; Wu, L.; Bai, X.; Zhai, S. Cytotoxicity-related bioeffects induced by nanoparticle: The role of surface chemistry. Front. Bioeng. Biotechnol. 2019, 7, 414. [Google Scholar] [CrossRef] [PubMed]







| Preparations | UV-Vis λmax (nm) | DLS | NTA | TEM (nm) | |||
|---|---|---|---|---|---|---|---|
| Z-Average Size (nm) | PDI | ζ-Potentials (mv) | Mean * (nm) | Mode * (nm) | |||
| Bare AuNPs (PEGylated) | 531.33 | 41.77 ± 1.26 | 0.315 | −35.53 ± 1.7 | 34.3 ± 0.2 | 34.7 ± 0.2 | 27.0 ± 3.0 |
| AuNP-NmAb Conjugates | 535.17 | 58.3 ± 0.36 | 0.264 | −0.048 ± 0.10 | 64.9 ± 2.9 | 71.4 ± 1.9 | 27.21 ± 3.08 |
| AuNP-NmAb +10% NaCl | 535.7 | 59.81 ± 1.18 | 0.277 | −10.2 ± 0.0 | - | - | - |
| AuNPs-NmAb +10% FBS | 536.17 | 73.17 ± 1.14 | 0.262 | −8.54 ± 1.33 | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anisuzzman, M.; Komalla, V.; Tarkistani, M.A.M.; Kayser, V. Anti-Tumor Activity of Novel Nimotuzumab-Functionalized Gold Nanoparticles as a Potential Immunotherapeutic Agent against Skin and Lung Cancers. J. Funct. Biomater. 2023, 14, 407. https://doi.org/10.3390/jfb14080407
Anisuzzman M, Komalla V, Tarkistani MAM, Kayser V. Anti-Tumor Activity of Novel Nimotuzumab-Functionalized Gold Nanoparticles as a Potential Immunotherapeutic Agent against Skin and Lung Cancers. Journal of Functional Biomaterials. 2023; 14(8):407. https://doi.org/10.3390/jfb14080407
Chicago/Turabian StyleAnisuzzman, Mohammad, Varsha Komalla, Mariam Abdulaziz M. Tarkistani, and Veysel Kayser. 2023. "Anti-Tumor Activity of Novel Nimotuzumab-Functionalized Gold Nanoparticles as a Potential Immunotherapeutic Agent against Skin and Lung Cancers" Journal of Functional Biomaterials 14, no. 8: 407. https://doi.org/10.3390/jfb14080407