Glutathione S-Transferases in Marine Copepods
Abstract
:1. Introduction
Glutathione S-Transferase Activity in Response to Environmental Stressors
| Species | GST | Stress | Response | Approach | Ref |
|---|---|---|---|---|---|
| Tigriopus japonicus | GST | Water-accommodated fraction (WAF) of crude oil | Increase | a | [11] |
| GST | WAF of crude oil | Increase | d | [12] | |
| GST | Endocrine disruptor chemicals (i.e., 4,4′-octylphenol and polychlorinated biphenyl) | Increase for 4,4′-octylphenol and decrease for polychlorinated biphenyl | a, c | [13] | |
| GST | Heavy metals (e.g., manganese, copper, nickel, cadmium, silver, and arsenic) | Increase | a, c, d | [10,14,15,16,17,50] | |
| GST | Brominated flame retardants (e.g., hexabromocyclododecanes) | Increase | a | [18] | |
| GST sigma, GST omega, GST mu, GST delta/epsilon | β-Naphtoflavone | Increase | a | [20] | |
| GST theta | Triclosan | Increase | a, d | [21] | |
| GST | Hydrogen peroxide | Increase | a, d | [14] | |
| Ten GSTs | Hydrogen peroxide and trace metals (i.e., silver, arsenic, cadmium, and copper) | GST sigma was the most up-regulated | a | [51] | |
| GST | Triphenyltin | Down | a | [23] | |
| GST | Cypermethrin | Increase | a, d | [22] | |
| GST | Multi-walled carbon nanotubes (MWCNTs) are nanoparticles widely applicable in various industrial fields. | Down | c, d | [24] | |
| GST | Low pH | Increase | a, d | [19] | |
| GST-kappa, GST-mu5, and GST-omega | Toxin-producing dinoflagellate Gymnodinium catenatum | Up | a | [40] | |
| GST | Two diatoms, Chaetoceros muelleri and Nitzschia closterium f. minutissima | No variation | d | [46] | |
| Tigriopus californicus | GSTs | Hydrogen peroxide and paraquat | Up/Down | b | [25] |
| GSTs | Hydrogen peroxide and yeast diet | Up only in males | b | [26] | |
| Paracyclopina nana | GST | Cypermethrin | Increase | a, d | [22] |
| GST | Copper | Increase | d | [10] | |
| GST | Methylmercury | Increase in activity, no variation in gene expression | a, c, d, | [36] | |
| GST | Polystyrene microbeads | Increase | d | [35] | |
| Eurytemora affinis | GST activity | Hydrophobic organic contaminant (HOC) | Increase | d | [27] |
| Calanus finmarchicus | GST | Naphtalene | Up | a | [29] |
| GST | Dispersed oil | Up | a | [30] | |
| GST | Water-accommodated fraction (WAF) of oil | Up | a | [11] | |
| GST | Mercury | Increase | a | [28] | |
| Three cytosolic GSTs and an omega GST | Alexandrium fundyense | Up | b | [39] | |
| Calanus glacialis | GST | Water-accommodated fraction (WAF) of oil | Up | a | [11] |
| GST | Mercury | Increase | a | [28] | |
| Calanus helgolandicus | GST | Skeletonema marinoi | Up-regulation for the Swedish copepod population | a | [42] |
| Microsomal GST3 | Skeletonema marinoi | Up | a, b | [41] | |
| GST | Sampling cruise in the Adriatic Sea (Mediterranean Sea) when a natural diatom bloom was observed | Up | a | [43] | |
| GST | Karenia brevis | Down | a | [44] | |
| Calanus sinicus | GST | Skeletonema marinoi | No variation | a | [45] |
| Calanus pacificus | GST | California West Coast Ocean Acidification Cruise Current System (effect of pH and temperature) | High GST activity | c, d | [49] |
| Centropages ponticus | GST | Cadmium chloride | Increase | d | [31] |
| Centropages tenuiremis | GST | Acidified seawater | No significant differences | c, d | [47] |
| Acartia tonsa | GST-theta and GST-mu | Polycyclic aromatic hydrocarbon (PAH) 1,2-dimethylnaphthalene | Increase | a | [32] |
| GST | Two-week field survey (effect of pH and temperature) | GST activity was higher in week II compared to week I, and, in particular, it was higher in the surface compared to the other depths for both weeks. | c, d | [48] | |
| Acartia bifilosa | GST | Two-week field survey (effect of pH and temperature) | GST activity was higher in week II compared to week I, and, in particular, it was higher in the surface compared to the other depths for both weeks. | c, d | [48] |
| Acartia pacifica | GST | Two diatoms, Chaetoceros muelleri and Nitzschia closterium f. minutissima | Increase with C. muelleri | d | [46] |
| Pseudodiaptomus poplesia | GST delta/epsilon and GST theta | PAH 1,2-dimethylnaphthalene, pyrene | GST delta/epsilon up-regulation for both stressors, GST theta only for 1,2-dimethylnaphthalene | a | [33] |
| Pseudodiaptomus annandalei | GST | Two diatoms, Chaetoceros muelleri and Nitzschia closterium f. minutissima | Increase with both algae | d | [46] |
| Eudiaptomus gracilis | GST | Ultraviolet radiation | Increase | d | [52] |
| Caligus rogercressereyi | GST | Delousing drug deltamethrin (AlphaMaxTM) | Down | b | [37] |
| Lepeophtheirus salmonis | Microsomal glutathione S-transferase 1 | Various lice stages from egg to adult lice | Constitutive transcript level throughout the studied development stages, with the highest levels in the ovaries or gut | a, b | [38] |
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lauritano, C.; Ianora, A. Chemical Defense in Marine Organisms. Mar. Drugs 2020, 18, 518. [Google Scholar] [CrossRef]
- Townsend, D.M.; Tew, K.D. The Role of Glutathione- S -Transferase in Anti-Cancer Drug Resistance. Oncogene 2003, 22, 7369–7375. [Google Scholar] [CrossRef] [Green Version]
- Lauritano, C.; Procaccini, G.; Ianora, A. Gene Expression Patterns and Stress Response in Marine Copepods. Mar. Environ. Res. 2012, 76, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Van Straalen, N.M.; Roelofs, D. An Introduction to Ecological Genomics, 2nd ed.; Oxford University Press: Oxford, UK; New York, NY, USA, 2012; ISBN 978-0-19-959468-9. [Google Scholar]
- Roncalli, V.; Cieslak, M.C.; Passamaneck, Y.; Christie, A.E.; Lenz, P.H. Glutathione S-Transferase (GST) Gene Diversity in the Crustacean Calanus finmarchicus—Contributors to Cellular Detoxification. PLoS ONE 2015, 10, e0123322. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.T. Zooplankton Fecal Pellets, Marine Snow, Phytodetritus and the Ocean’s Biological Pump. Prog. Oceanogr. 2015, 130, 205–248. [Google Scholar] [CrossRef]
- Carotenuto, Y.; Vitiello, V.; Gallo, A.; Libralato, G.; Trifuoggi, M.; Toscanesi, M.; Lofrano, G.; Esposito, F.; Buttino, I. Assessment of the Relative Sensitivity of the Copepods Acartia tonsa and Acartia clausi Exposed to Sediment-Derived Elutriates from the Bagnoli-Coroglio Industrial Area. Mar. Environ. Res. 2020, 155, 104878. [Google Scholar] [CrossRef]
- Russo, E.; Ianora, A.; Carotenuto, Y. Re-Shaping Marine Plankton Communities: Effects of Diatom Oxylipins on Copepods and Beyond. Mar. Biol. 2018, 166, 9. [Google Scholar] [CrossRef]
- Choi, B.-S.; Kim, D.-H.; Kim, M.-S.; Park, J.C.; Lee, Y.H.; Kim, H.-J.; Jeong, C.-B.; Hagiwara, A.; Souissi, S.; Lee, J.-S. The Genome of the European Estuarine Calanoid Copepod Eurytemora affinis: Potential Use in Molecular Ecotoxicology. Mar. Pollut. Bull. 2021, 166, 112190. [Google Scholar] [CrossRef]
- Park, J.C.; Lee, M.-C.; Yoon, D.-S.; Han, J.; Park, H.G.; Hwang, U.-K.; Lee, J.-S. Genome-Wide Identification and Expression of the Entire 52 Glutathione S-Transferase (GST) Subfamily Genes in the Cu2+-Exposed Marine Copepods Tigriopus japonicus and Paracyclopina nana. Aquat. Toxicol. 2019, 209, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.H.; Altin, D.; Rørvik, S.F.; Øverjordet, I.B.; Olsen, A.J.; Nordtug, T. Comparative Study on Acute Effects of Water Accommodated Fractions of an Artificially Weathered Crude Oil on Calanus finmarchicus and Calanus glacialis (Crustacea: Copepoda). Sci. Total Environ. 2011, 409, 704–709. [Google Scholar] [CrossRef]
- Han, J.; Won, E.-J.; Hwang, D.-S.; Shin, K.-H.; Lee, Y.S.; Leung, K.M.-Y.; Lee, S.-J.; Lee, J.-S. Crude Oil Exposure Results in Oxidative Stress-Mediated Dysfunctional Development and Reproduction in the Copepod Tigriopus japonicus and Modulates Expression of Cytochrome P450 (CYP) Genes. Aquat. Toxicol. 2014, 152, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-M.; Park, T.-J.; Jung, S.-O.; Seo, J.S.; Park, H.G.; Hagiwara, A.; Yoon, Y.-D.; Lee, J.-S. Cloning and Characterization of Glutathione S-Transferase Gene in the Intertidal Copepod Tigriopus japonicus and Its Expression after Exposure to Endocrine-Disrupting Chemicals. Mar. Environ. Res. 2006, 62, S219–S223. [Google Scholar] [CrossRef]
- Lee, Y.-M.; Lee, K.-W.; Park, H.; Park, H.G.; Raisuddin, S.; Ahn, I.-Y.; Lee, J.-S. Sequence, Biochemical Characteristics and Expression of a Novel Sigma-Class of Glutathione S-Transferase from the Intertidal Copepod, Tigriopus japonicus with a Possible Role in Antioxidant Defense. Chemosphere 2007, 69, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Rhee, J.-S.; Kim, B.-M.; Jeong, C.-B.; Leung, K.M.Y.; Park, G.S.; Lee, J.-S. Development of Enzyme-Linked Immunosorbent Assay (ELISA) for Glutathione S-Transferase (GST-S) Protein in the Intertidal Copepod Tigriopus japonicus and Its Application for Environmental Monitoring. Chemosphere 2013, 93, 2458–2466. [Google Scholar] [CrossRef]
- Wang, M.; Wang, G. Oxidative Damage Effects in the Copepod Tigriopus japonicus Mori Experimentally Exposed to Nickel. Ecotoxicology 2010, 19, 273–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.-H.; Wang, G.-Z. Biochemical Response of the Copepod Tigriopus japonicus Mori Experimentally Exposed to Cadmium. Arch. Environ. Contam. Toxicol. 2009, 57, 707–717. [Google Scholar] [CrossRef] [Green Version]
- Shi, D.; Lv, D.; Liu, W.; Shen, R.; Li, D.; Hong, H. Accumulation and Developmental Toxicity of Hexabromocyclododecanes (HBCDs) on the Marine Copepod Tigriopus japonicus. Chemosphere 2017, 167, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Kang, H.-M.; Kim, M.-S.; Wang, M.; Kim, J.H.; Jeong, C.-B.; Lee, J.-S. Effects of Ocean Acidification on Life Parameters and Antioxidant System in the Marine Copepod Tigriopus japonicus. Aquat. Toxicol. 2019, 212, 186–193. [Google Scholar] [CrossRef]
- Rhee, J.-S.; Lee, Y.-M.; Kim, B.-M.; Leung, K.M.Y.; Kim, I.-C.; Yim, J.H.; Lee, J.-S. β -Naphthoflavone Induces Oxidative Stress in the Intertidal Copepod, Tigriopus japonicus: β-NF-Induced Oxidative Stress in Copepod. Environ. Toxicol. 2015, 30, 332–342. [Google Scholar] [CrossRef]
- Park, J.C.; Han, J.; Lee, M.-C.; Seo, J.S.; Lee, J.-S. Effects of Triclosan (TCS) on Fecundity, the Antioxidant System, and Oxidative Stress-Mediated Gene Expression in the Copepod Tigriopus japonicus. Aquat. Toxicol. 2017, 189, 16–24. [Google Scholar] [CrossRef]
- Zhou, J.; Kang, H.-M.; Lee, Y.H.; Jeong, C.-B.; Park, J.C.; Lee, J.-S. Adverse Effects of a Synthetic Pyrethroid Insecticide Cypermethrin on Life Parameters and Antioxidant Responses in the Marine Copepods Paracyclopina nana and Tigriopus japonicus. Chemosphere 2019, 217, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Yi, A.X.; Han, J.; Lee, J.-S.; Leung, K.M.Y. Ecotoxicity of Triphenyltin on the Marine Copepod Tigriopus japonicus at Various Biological Organisations: From Molecular to Population-Level Effects. Ecotoxicology 2014, 23, 1314–1325. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Won, E.-J.; Kang, H.-M.; Hwang, D.-S.; Kim, D.-H.; Kim, R.-K.; Lee, S.-J.; Lee, J.-S. Effects of Multi-Walled Carbon Nanotube (MWCNT) on Antioxidant Depletion, the ERK Signaling Pathway, and Copper Bioavailability in the Copepod (Tigriopus japonicus). Aquat. Toxicol. 2016, 171, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Flanagan, B.A.; Partridge, M.; Huang, E.J.; Edmands, S. Sex Differences in Early Transcriptomic Responses to Oxidative Stress in the Copepod Tigriopus californicus. BMC Genom. 2020, 21, 759. [Google Scholar] [CrossRef]
- Li, N.; Arief, N.; Edmands, S. Effects of Oxidative Stress on Sex-Specific Gene Expression in the Copepod Tigriopus californicus Revealed by Single Individual RNA-Seq. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 31, 100608. [Google Scholar] [CrossRef] [PubMed]
- Cailleaud, K.; Forget-Leray, J.; Peluhet, L.; LeMenach, K.; Souissi, S.; Budzinski, H. Tidal Influence on the Distribution of Hydrophobic Organic Contaminants in the Seine Estuary and Biomarker Responses on the Copepod Eurytemora affinis. Environ. Pollut. 2009, 157, 64–71. [Google Scholar] [CrossRef]
- Øverjordet, I.B.; Altin, D.; Berg, T.; Jenssen, B.M.; Gabrielsen, G.W.; Hansen, B.H. Acute and Sub-Lethal Response to Mercury in Arctic and Boreal Calanoid Copepods. Aquat. Toxicol. 2014, 155, 160–165. [Google Scholar] [CrossRef]
- Hansen, B.H.; Altin, D.; Vang, S.-H.; Nordtug, T.; Olsen, A.J. Effects of Naphthalene on Gene Transcription in Calanus finmarchicus (Crustacea: Copepoda). Aquat. Toxicol. 2008, 86, 157–165. [Google Scholar] [CrossRef]
- Hansen, B.H.; Nordtug, T.; Altin, D.; Booth, A.; Hessen, K.M.; Olsen, A.J. Gene Expression of GST and CYP330A1 in Lipid-Rich and Lipid-Poor Female Calanus finmarchicus (Copepoda: Crustacea) Exposed to Dispersed Oil. J. Toxicol. Environ. Health A 2009, 72, 131–139. [Google Scholar] [CrossRef]
- Ensibi, C.; Yahia, M.N.D. Toxicity Assessment of Cadmium Chloride on Planktonic Copepods Centropages ponticus Using Biochemical Markers. Toxicol. Rep. 2017, 4, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, B.; Zeng, S.; Gong, Z.; Jing, F.; Zhang, J. Glutathione S-Transferase (GST) Genes from Marine Copepods Acartia tonsa: CDNA Cloning and MRNA Expression in Response to 1,2-Dimethylnaphthalene. Aquat. Toxicol. 2020, 224, 105480. [Google Scholar] [CrossRef]
- Zhuang, Y.; Yang, F.; Xu, D.; Chen, H.; Zhang, H.; Liu, G. Spliced Leader-Based Analyses Reveal the Effects of Polycyclic Aromatic Hydrocarbons on Gene Expression in the Copepod Pseudodiaptomus poplesia. Aquat. Toxicol. 2017, 183, 114–126. [Google Scholar] [CrossRef]
- Beiras, R.; Tato, T.; López-Ibáñez, S. A 2-Tier Standard Method to Test the Toxicity of Microplastics in Marine Water Using Paracentrotus lividus and Acartia clausi Larvae. Environ. Toxicol. Chem. 2019, 38, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Jeong, C.-B.; Kang, H.-M.; Lee, M.-C.; Kim, D.-H.; Han, J.; Hwang, D.-S.; Souissi, S.; Lee, S.-J.; Shin, K.-H.; Park, H.G.; et al. Adverse Effects of Microplastics and Oxidative Stress-Induced MAPK/Nrf2 Pathway-Mediated Defense Mechanisms in the Marine Copepod Paracyclopina nana. Sci. Rep. 2017, 7, 41323. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.H.; Kim, D.-H.; Kang, H.-M.; Wang, M.; Jeong, C.-B.; Lee, J.-S. Adverse Effects of Methylmercury (MeHg) on Life Parameters, Antioxidant Systems, and MAPK Signaling Pathways in the Rotifer Brachionus koreanus and the Copepod Paracyclopina nana. Aquat. Toxicol. 2017, 190, 181–189. [Google Scholar] [CrossRef]
- Chávez-Mardones, J.; Gallardo-Escárate, C. Next-Generation Transcriptome Profiling of the Salmon Louse Caligus rogercresseyi Exposed to Deltamethrin (AlphaMaxTM): Discovery of Relevant Genes and Sex-Related Differences. Mar. Biotechnol. 2015, 17, 793–810. [Google Scholar] [CrossRef]
- Dalvin, S.; Eichner, C.; Dondrup, M.; Øvergård, A.-C. Roles of Three Putative Salmon Louse (Lepeophtheirus salmonis) Prostaglandin E2 Synthases in Physiology and Host–Parasite Interactions. Parasites Vectors 2021, 14, 206. [Google Scholar] [CrossRef]
- Roncalli, V.; Turner, J.T.; Kulis, D.; Anderson, D.M.; Lenz, P.H. The Effect of the Toxic Dinoflagellate Alexandrium fundyense on the Fitness of the Calanoid Copepod Calanus finmarchicus. Harmful Algae 2016, 51, 56–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Park, J.S.; Park, Y.; Lee, J.; Shin, H.H.; Lee, K.-W. Effects of Paralytic Shellfish Poisoning Toxin-Producing Dinoflagellate Gymnodinium catenatum on the Marine Copepod Tigriopus japonicus. Mar. Pollut. Bull. 2021, 163, 111937. [Google Scholar] [CrossRef] [PubMed]
- Asai, S.; Sanges, R.; Lauritano, C.; Lindeque, P.K.; Esposito, F.; Ianora, A.; Carotenuto, Y. De Novo Transcriptome Assembly and Gene Expression Profiling of the Copepod Calanus helgolandicus Feeding on the PUA-Producing Diatom Skeletonema Marinoi. Mar. Drugs 2020, 18, 392. [Google Scholar] [CrossRef]
- Lauritano, C.; Carotenuto, Y.; Miralto, A.; Procaccini, G.; Ianora, A. Copepod Population-Specific Response to a Toxic Diatom Diet. PLoS ONE 2012, 7, e47262. [Google Scholar] [CrossRef] [Green Version]
- Lauritano, C.; Romano, G.; Roncalli, V.; Amoresano, A.; Fontanarosa, C.; Bastianini, M.; Braga, F.; Carotenuto, Y.; Ianora, A. New Oxylipins Produced at the End of a Diatom Bloom and Their Effects on Copepod Reproductive Success and Gene Expression Levels. Harmful Algae 2016, 55, 221–229. [Google Scholar] [CrossRef]
- Lauritano, C.; Carotenuto, Y.; Procaccini, G.; Turner, J.T.; Ianora, A. Changes in Expression of Stress Genes in Copepods Feeding upon a Non-Brevetoxin-Producing Strain of the Dinoflagellate Karenia brevis. Harmful Algae 2013, 28, 23–30. [Google Scholar] [CrossRef]
- Lauritano, C.; Carotenuto, Y.; Vitiello, V.; Buttino, I.; Romano, G.; Hwang, J.-S.; Ianora, A. Effects of the Oxylipin-Producing Diatom Skeletonema marinoi on Gene Expression Levels of the Calanoid Copepod Calanus sinicus. Mar. Genom. 2015, 24, 89–94. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, G.; Zeng, C.; Wu, L. Comparative Study on the Effects of Two Diatoms as Diets on Planktonic Calanoid and Benthic Harpacticoid Copepods. J. Exp. Zool. Part Ecol. Integr. Physiol. 2018, 329, 140–148. [Google Scholar] [CrossRef]
- Zhang, D.; Li, S.; Wang, G.; Guo, D.; Xing, K.; Zhang, S. Biochemical Responses of the Copepod Centropages tenuiremis to CO2-Driven Acidified Seawater. Water Sci. Technol. 2012, 65, 30–37. [Google Scholar] [CrossRef]
- Glippa, O.; Engström-Öst, J.; Kanerva, M.; Rein, A.; Vuori, K. Oxidative Stress and Antioxidant Defense Responses in Acartia Copepods in Relation to Environmental Factors. PLoS ONE 2018, 13, e0195981. [Google Scholar] [CrossRef] [Green Version]
- Engström-Öst, J.; Glippa, O.; Feely, R.A.; Kanerva, M.; Keister, J.E.; Alin, S.R.; Carter, B.R.; McLaskey, A.K.; Vuori, K.A.; Bednaršek, N. Eco-Physiological Responses of Copepods and Pteropods to Ocean Warming and Acidification. Sci. Rep. 2019, 9, 4748. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-M.; Choi, B.-S.; Lee, K.-W.; Ki, J.-S.; Kim, I.-C.; Choi, I.-Y.; Rhee, J.-S.; Lee, J.-S. Expression Profile Analysis of Antioxidative Stress and Developmental Pathway Genes in the Manganese-Exposed Intertidal Copepod Tigriopus japonicus with 6K Oligochip. Chemosphere 2013, 92, 1214–1223. [Google Scholar] [CrossRef] [PubMed]
- Lee, K. Expression of Glutathione S-Transferase (GST) Genes in the Marine Copepod Tigriopus japonicus Exposed to Trace Metals. Aquat. Toxicol. 2008, 89, 158–166. [Google Scholar] [CrossRef]
- Souza, M.S.; Hansson, L.-A.; Hylander, S.; Modenutti, B.; Balseiro, E. Rapid Enzymatic Response to Compensate UV Radiation in Copepods. PLoS ONE 2012, 7, e32046. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Apple, R.D.; Bairoch, A. ExPASy: The Proteomics Server for in-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Park, J.C.; Hagiwara, A.; Park, H.G.; Lee, J.-S. The Glutathione S-Transferase Genes in Marine Rotifers and Copepods: Identification of GSTs and Applications for Ecotoxicological Studies. Mar. Pollut. Bull. 2020, 156, 111080. [Google Scholar] [CrossRef] [PubMed]
- Jeong, C.-B.; Lee, B.-Y.; Choi, B.-S.; Kim, M.-S.; Park, J.C.; Kim, D.-H.; Wang, M.; Park, H.G.; Lee, J.-S. The Genome of the Harpacticoid Copepod Tigriopus japonicus: Potential for Its Use in Marine Molecular Ecotoxicology. Aquat. Toxicol. 2020, 222, 105462. [Google Scholar] [CrossRef]
- Lauritano, C.; Borra, M.; Carotenuto, Y.; Biffali, E.; Miralto, A.; Procaccini, G.; Ianora, A. Molecular Evidence of the Toxic Effects of Diatom Diets on Gene Expression Patterns in Copepods. PLoS ONE 2011, 6, e26850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauritano, C.; Borra, M.; Carotenuto, Y.; Biffali, E.; Miralto, A.; Procaccini, G.; Ianora, A. First Molecular Evidence of Diatom Effects in the Copepod Calanus helgolandicus. J. Exp. Mar. Biol. Ecol. 2011, 404, 79–86. [Google Scholar] [CrossRef]
- Carotenuto, Y.; Esposito, F.; Pisano, F.; Lauritano, C.; Perna, M.; Miralto, A.; Ianora, A. Multi-Generation Cultivation of the Copepod Calanus helgolandicus in a Re-Circulating System. J. Exp. Mar. Biol. Ecol. 2012, 418–419, 46–58. [Google Scholar] [CrossRef]
- Ianora, A.; Miralto, A. Toxigenic Effects of Diatoms on Grazers, Phytoplankton and Other Microbes: A Review. Ecotoxicology 2010, 19, 493–511. [Google Scholar] [CrossRef] [PubMed]
- Carotenuto, Y.; Dattolo, E.; Lauritano, C.; Pisano, F.; Sanges, R.; Miralto, A.; Procaccini, G.; Ianora, A. Insights into the Transcriptome of the Marine Copepod Calanus helgolandicus Feeding on the Oxylipin-Producing Diatom Skeletonema marinoi. Harmful Algae 2014, 31, 153–162. [Google Scholar] [CrossRef] [PubMed]
- da Fonseca, R.R.; Johnson, W.E.; O’Brien, S.J.; Vasconcelos, V.; Antunes, A. Molecular Evolution and the Role of Oxidative Stress in the Expansion and Functional Diversification of Cytosolic Glutathione Transferases. BMC Evol. Biol. 2010, 10, 281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Favreau, L.V.; Pickett, C.B. Transcriptional Regulation of the Rat NAD(P)H:Quinone Reductase Gene. Identification of Regulatory Elements Controlling Basal Level Expression and Inducible Expression by Planar Aromatic Compounds and Phenolic Antioxidants. J. Biol. Chem. 1991, 266, 4556–4561. [Google Scholar] [CrossRef]
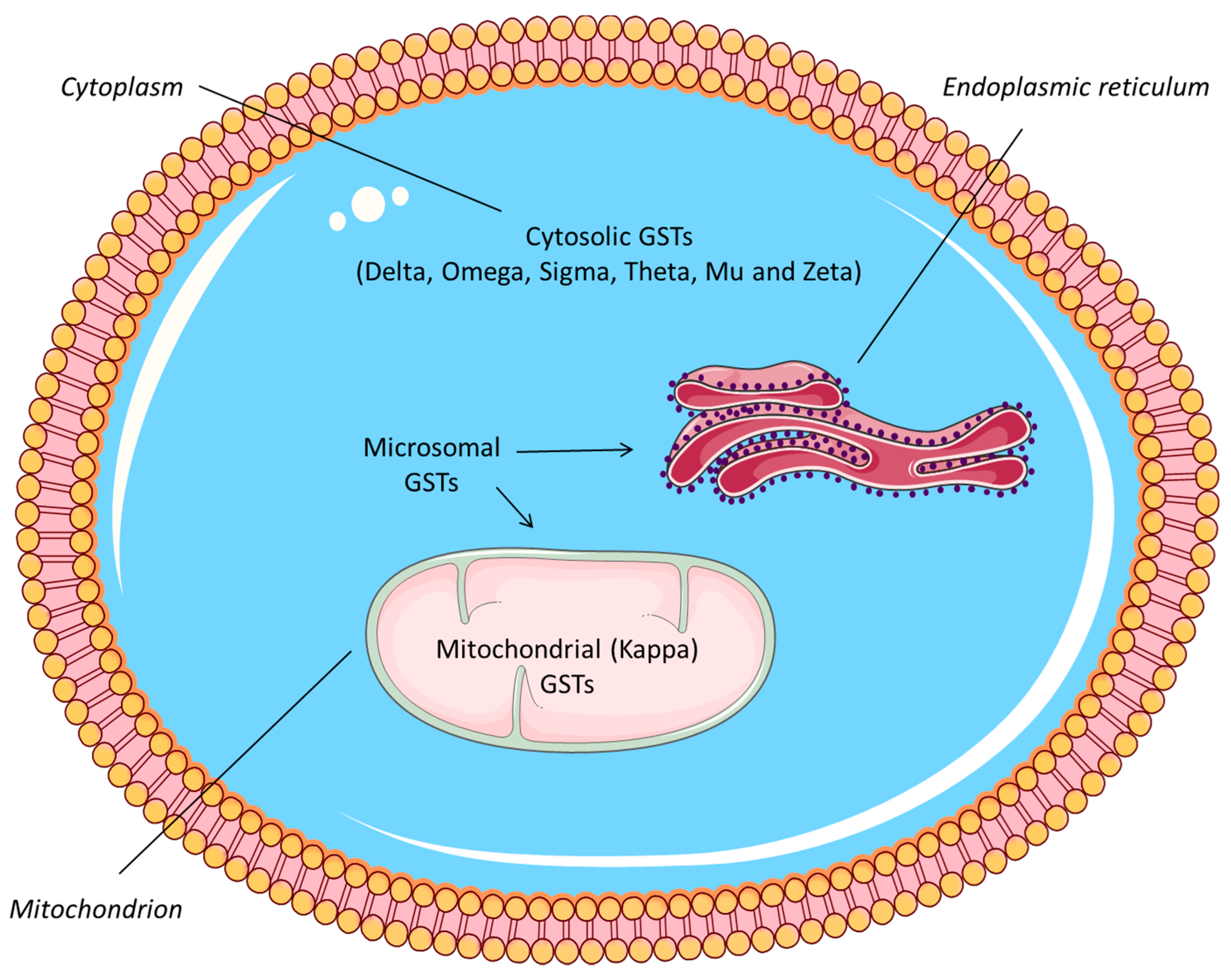
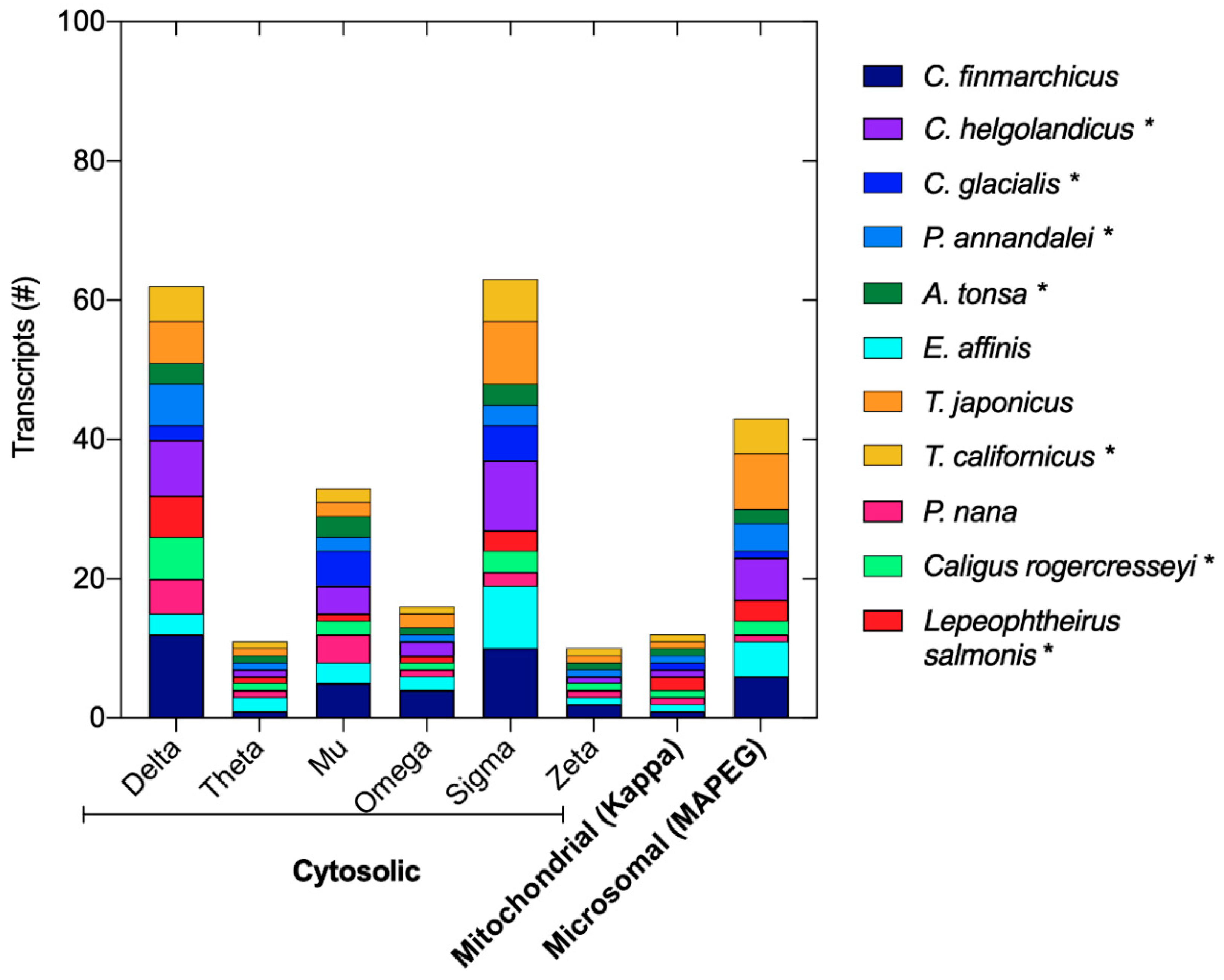
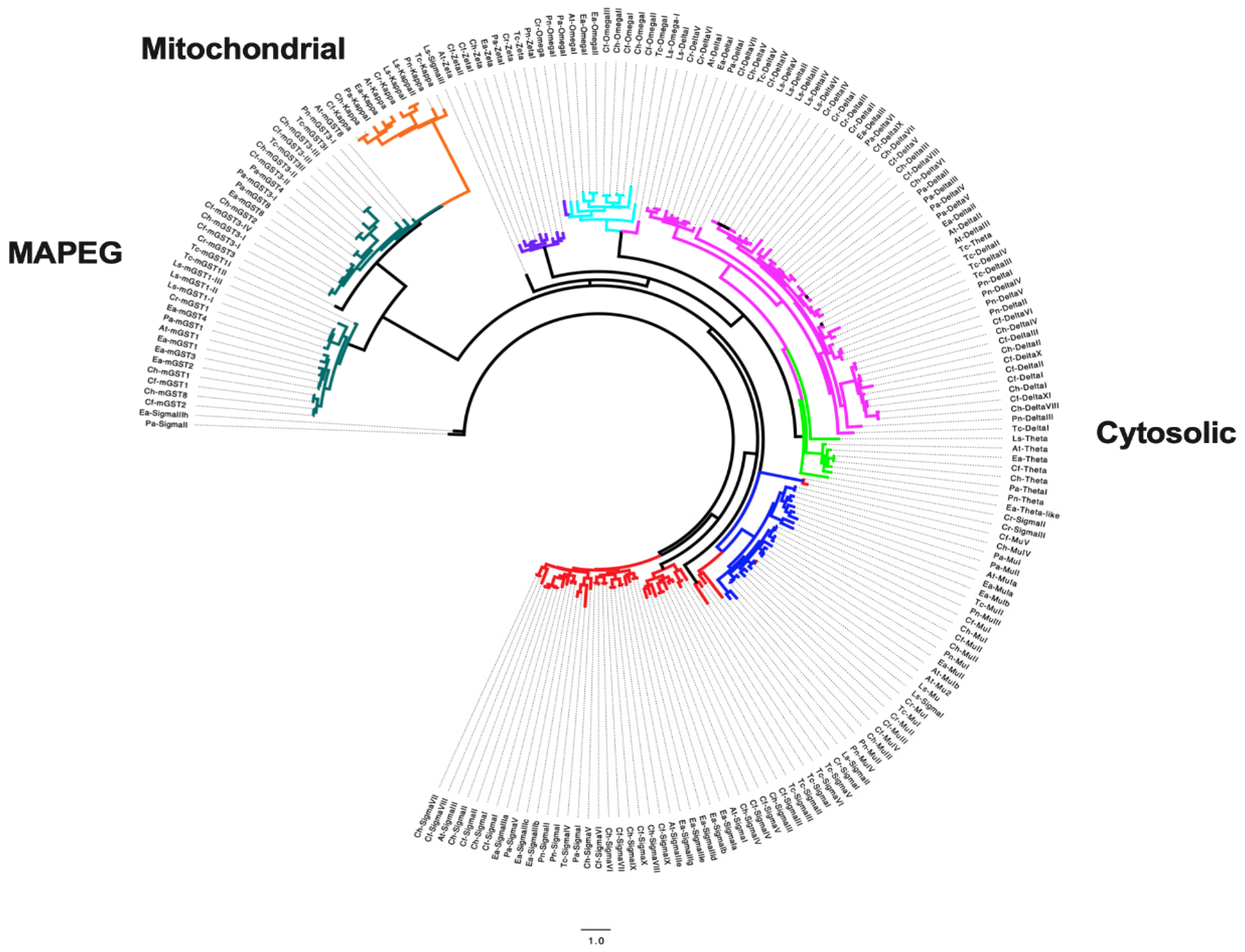
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lauritano, C.; Carotenuto, Y.; Roncalli, V. Glutathione S-Transferases in Marine Copepods. J. Mar. Sci. Eng. 2021, 9, 1025. https://doi.org/10.3390/jmse9091025
Lauritano C, Carotenuto Y, Roncalli V. Glutathione S-Transferases in Marine Copepods. Journal of Marine Science and Engineering. 2021; 9(9):1025. https://doi.org/10.3390/jmse9091025
Chicago/Turabian StyleLauritano, Chiara, Ylenia Carotenuto, and Vittoria Roncalli. 2021. "Glutathione S-Transferases in Marine Copepods" Journal of Marine Science and Engineering 9, no. 9: 1025. https://doi.org/10.3390/jmse9091025








