Transcriptome Analysis Reveals the Endocrine Regulation on the Expression of IAG in Litopenaeus vannamei
Abstract
:1. Introduction
2. Results
2.1. Transcriptome Data Mapping and Analysis
2.2. Identification of Differentially Expressed Genes (DEGs)
2.3. Identification of DEGs Related to Male Differentiation
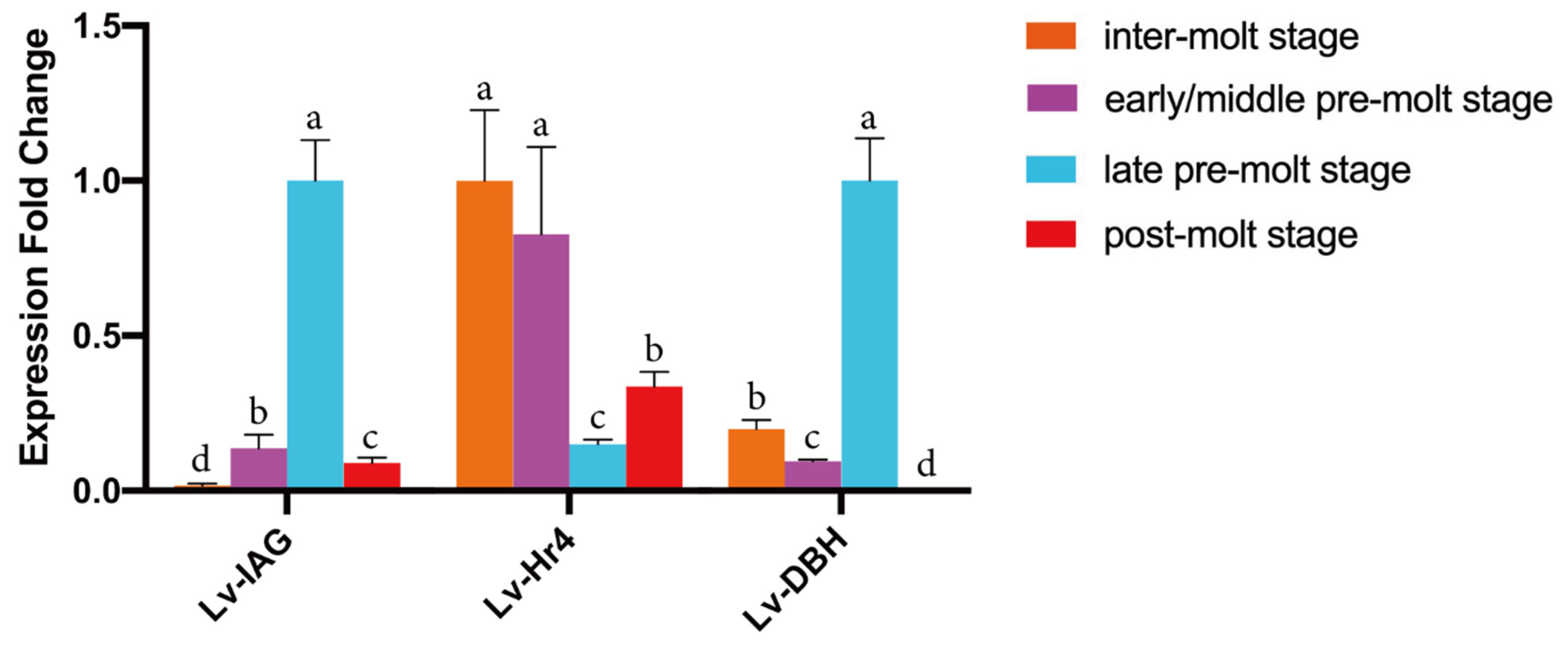
2.4. Expression Profiles of Selected Genes at Different Molt Stages
2.5. Lv-IAG Expression Profile in Molting Cycle with 20E Treatment
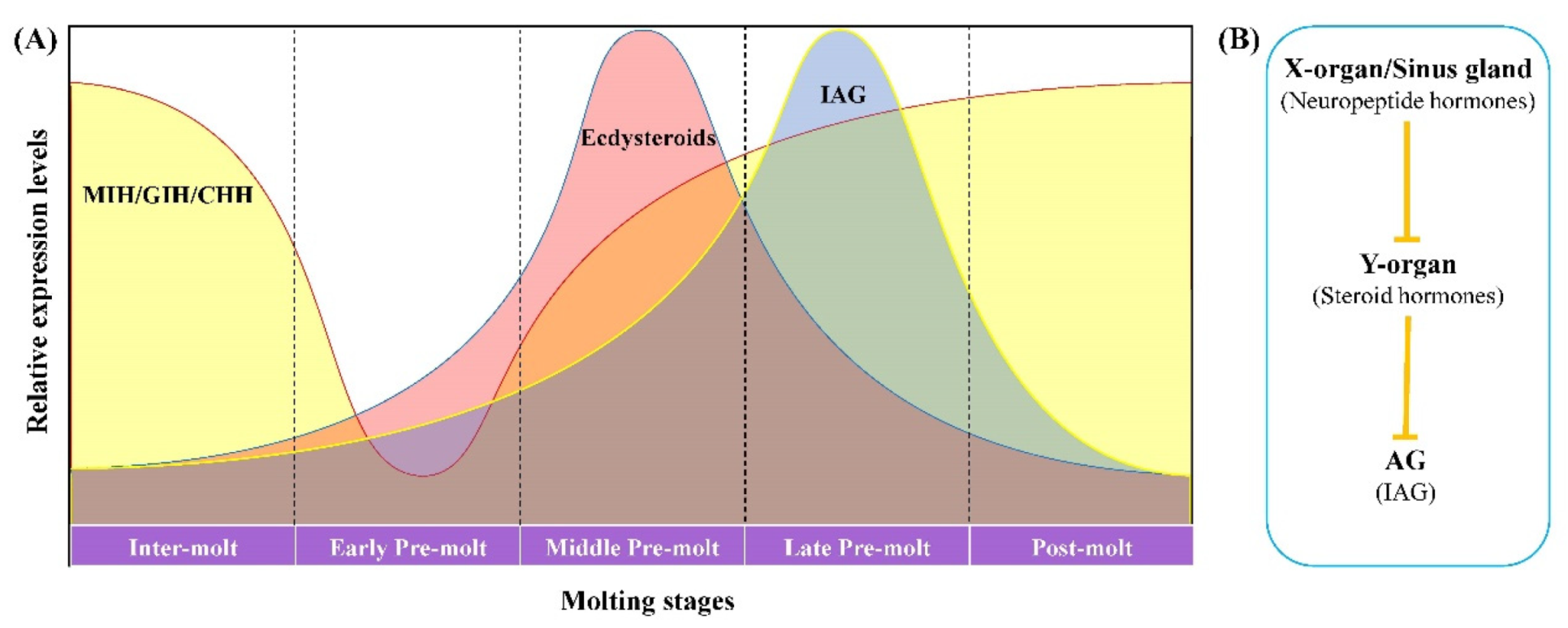
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. Unilateral Eyestalk Ablation
4.3. Total RNA Preparation and cDNA Synthesis
4.4. RNA-Sequencing
4.5. Bioinformatic Analyses and DEGs Identification
4.6. AG Collection at Different Molting Stages
4.7. 20-Hydroxyecdysone (20E) Injection
4.8. RT-qPCR Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sagi, A.; Aflalo, E.D. The androgenic gland and monosex culture of freshwater prawn Macrobrachium rosenbergii (De Man): A biotechnological perspective. Aquac. Res. 2005, 36, 231–237. [Google Scholar] [CrossRef]
- Laramore, S.; Laramore, C.R.; ScarpaEffect, J. Effect of low salinity on growth and survival of postlarvae and juvenile Litopenaeus vannamei. J. World Aquac. Soc. 2001, 32, 385–390. [Google Scholar] [CrossRef]
- Gitterle, T.; Rye, M.; Salte, R.; Cock, J.; Johansen, H.; Lozano, C.; Arturo Suárez, J.; Gjerde, B. Genetic (co)variation in harvest body weight and survival in Penaeus (Litopenaeus) vannamei under standard commercial conditions. Aquaculture 2005, 243, 83–92. [Google Scholar] [CrossRef]
- Levy, T.; Sagi, A. The “IAG-Switch”—A key controlling element in decapod crustacean sex differentiation. Front. Endocrinol. 2020, 11, 651. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Rosen, O.; Sagi, A. Timing sexual differentiation: Full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, Macrobrachium rosenbergii. Biol. Reprod. 2012, 86, 90. [Google Scholar] [CrossRef]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Raviv, S.; Glazer, L.; Sagi, A. Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology 2009, 150, 1278–1286. [Google Scholar] [CrossRef] [PubMed]
- Rosen, O.; Manor, R.; Weil, S.; Gafni, O.; Linial, A.; Aflalo, E.D.; Ventura, T.; Sagi, A. A sexual shift induced by silencing of a single insulin-like gene in crayfish: Ovarian upregulation and testicular degeneration. PLoS ONE 2010, 5, e15281. [Google Scholar] [CrossRef] [PubMed]
- Manor, R.; Weil, S.; Oren, S.; Glazer, L.; Aflalo, E.D.; Ventura, T.; Chalifa-Caspi, V.; Lapidot, M.; Sagi, A. Insulin and gender: An insulin-like gene expressed exclusively in the androgenic gland of the male crayfish. Gen. Comp. Endocrinol. 2007, 150, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Sagi, A. The insulin-like androgenic gland hormone in crustaceans: From a single gene silencing to a wide array of sexual manipulation-based biotechnologies. Biotechnol. Adv. 2012, 30, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Khalaila, I.; Katz, T.; Abdu, U.; Yehezkel, G.; Sagi, A. Effects of implantation of hypertrophied androgenic glands on sexual characters and physiology of the reproductive system in the female red claw crayfish, Cherax quadricarinatus. Gen. Comp. Endocrinol. 2001, 121, 242–249. [Google Scholar] [CrossRef]
- Katakura, Y. Masculinization of females of the isopod crustacean, Armadillidium vulgare, following injections of an active extract of the androgenic gland. Gen. Comp. Endocrinol. 1983, 49, 57–62. [Google Scholar] [CrossRef]
- Yu, Y.-Q.; Ma, W.-M.; Zeng, Q.-G.; Qian, Y.-Q.; Yang, J.-S.; Yang, W.-J. Molecular cloning and sexually dimorphic expression of two dmrt genes in the giant freshwater prawn, Macrobrachium rosenbergii. Agric. Res. 2014, 3, 181–191. [Google Scholar] [CrossRef]
- Zheng, J.; Cheng, S.; Jia, Y.; Gu, Z.; Li, F.; Chi, M.; Liu, S.; Jiang, W. Molecular identification and expression profiles of four splice variants of sex-lethal gene in Cherax quadricarinatus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2019, 234, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, F.; Yu, K.; Xiang, J. Identification and characterization of a doublesex gene which regulates the expression of insulin-like androgenic gland hormone in Fenneropenaeus chinensis. Gene 2018, 649, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Khalaila, I.; Manor, R.; Weil, S.; Granot, Y.; Keller, R.; Sagi, A. The eyestalk-androgenic gland-testis endocrine axis in the crayfish Cherax quadricarinatus. Gen. Comp. Endocrinol. 2002, 127, 147–156. [Google Scholar] [CrossRef]
- Foulks, N.B.; Hoffman, D.L. The effects of eyestalk ablation and B-ecdysone on RNA synthesis in the androgenic glands of the protandric shrimp, Pandalus platyceros brandt. Gen. Comp. Endocrinol. 1974, 22, 439–447. [Google Scholar] [CrossRef]
- Sroyraya, M.; Chotwiwatthanakun, C.; Stewart, M.J.; Soonklang, N.; Kornthong, N.; Phoungpetchara, I.; Hanna, P.J.; Sobhon, P. Bilateral eyestalk ablation of the blue swimmer crab, Portunus pelagicus, produces hypertrophy of the androgenic gland and an increase of cells producing insulin-like androgenic gland hormone. Tissue Cell 2010, 42, 293–300. [Google Scholar] [CrossRef]
- Phoungpetchara, I.; Tinikul, Y.; Poljaroen, J.; Chotwiwatthanakun, C.; Vanichviriyakit, R.; Sroyraya, M.; Hanna, P.J.; Sobhon, P. Cells producing insulin-like androgenic gland hormone of the giant freshwater prawn, Macrobrachium rosenbergii, proliferate following bilateral eyestalk-ablation. Tissue Cell 2011, 43, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Grecia, V.-I.; Danitzia, A.G.-T.; Rodolfo, G.-T.; Píndaro, Á.-R.; Humberto, M.-R.; Rafael, C.-R. Quantitative analysis of hypertrophy and hyperactivity in the androgenic gland of eyestalk-ablated male Pacific white shrimp Litopenaeus vannamei during molt stages. Aquaculture 2015, 439, 7–13. [Google Scholar] [CrossRef]
- Chung, J.S.; Manor, R.; Sagi, A. Cloning of an insulin-like androgenic gland factor (IAG) from the blue crab, Callinectes sapidus: Implications for eyestalk regulation of IAG expression. Gen. Comp. Endocrinol. 2011, 173, 4–10. [Google Scholar] [CrossRef]
- Hopkins, P.M. The eyes have it: A brief history of crustacean neuroendocrinology. Gen. Comp. Endocrinol. 2012, 175, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Lachaise, F.; Le Roux, A.; Hubert, M.; Lafont, R. The molting gland of crustaceans: Localization, activity, and endocrine control (A review). J. Crustacean Biol. 1993, 13, 198–234. [Google Scholar] [CrossRef]
- Guo, Q.; Li, S.; Lv, X.; Xiang, J.; Manor, R.; Sagi, A.; Li, F. Sex-biased CHHs and their putative receptor regulate the expression of IAG gene in the shrimp Litopenaeus vannamei. Front. Physiol. 2019, 10, 1525. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, C.; Greve, P.; Martin, G. Overview on the sub-grouping of the crustacean hyperglycemic hormone family. Neuropeptides 1999, 33, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Liu, J.; Liu, F.; Huang, Y.; Wang, G.; Ye, H. Crustacean female sex hormone from the mud crab Scylla paramamosain is highly expressed in prepubertal males and inhibits the development of androgenic gland. Front. Physiol. 2018, 9, 924. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Lu, B.; Wang, G.; Ye, H. Transcriptional Inhibition of Sp-IAG by Crustacean Female Sex Hormone in the Mud Crab, Scylla paramamosain. Int. J. Mol. Sci. 2020, 21, 5300. [Google Scholar] [CrossRef]
- Li, F.; Bai, H.; Zhang, W.; Fu, H.; Jiang, F.; Liang, G.; Jin, S.; Sun, S.; Qiao, H. Cloning of genomic sequences of three crustacean hyperglycemic hormone superfamily genes and elucidation of their roles of regulating insulin-like androgenic gland hormone gene. Gene 2015, 561, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Jun-Xia, C.; Guo-Li, Y.; Wei-Jun, Y. Identification of a novel male reproduction-related gene and its regulated expression patterns in the prawn, Macrobrachium rosenbergii. Peptides 2006, 27, 728–735. [Google Scholar] [CrossRef]
- Sroyraya, M.; Hanna, P.J.; Changklungmoa, N.; Senarai, T.; Siangcham, T.; Tinikul, Y.; Sobhon, P. Expression of the male reproduction-related gene in spermatic ducts of the blue swimming crab, Portunus pelagicus, and transfer of modified protein to the sperm acrosome. Microsc. Res. Tech. 2013, 76, 102–112. [Google Scholar] [CrossRef]
- Uchimura, T.; Hara, S.; Yazawa, T.; Kamei, Y.; Kitano, T. Involvement of heat shock proteins on the transcriptional regulation of corticotropin-releasing hormone in medaka. Front. Endocrinol. 2019, 10, 529. [Google Scholar] [CrossRef]
- Chandler, J.C.; Aizen, J.; Elizur, A.; Battaglene, S.C.; Ventura, T. Male sexual development and the androgenic gland: Novel insights through the de novo assembled transcriptome of the eastern spiny lobster, Sagmariasus verreauxi. Sex. Dev. 2015, 9, 338–354. [Google Scholar] [CrossRef]
- Fernandez, I.; Vijayakumar, P.; Marques, C.; Cancela, M.L.; Gavaia, P.J.; Laize, V. Zebrafish vitamin K epoxide reductases: Expression in vivo, along extracellular matrix mineralization and under phylloquinone and warfarin in vitro exposure. Fish. Physiol. Biochem. 2015, 41, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Beato, S.; Toledo-Solis, F.J.; Fernandez, I. Vitamin K in vertebrates’ reproduction: Further puzzling pieces of evidence from teleost fish species. Biomolecules 2020, 10, 1303. [Google Scholar] [CrossRef]
- Ma, H.; Zhang, B.L.; Liu, B.Y.; Shi, S.; Gao, D.Y.; Zhang, T.C.; Shi, H.J.; Li, Z.; Shum, W.W. Vitamin K2-dependent GGCX and MGP are required for homeostatic calcium regulation of sperm maturation. iScience 2019, 14, 210–225. [Google Scholar] [CrossRef]
- Beckstead, R.B.; Lam, G.; Thummel, C.S. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome. Biol. 2005, 6, R99. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.Y.; Meng, Q.W.; Deng, P.; Guo, W.C.; Li, G.Q. Leptinotarsa hormone receptor 4 (HR4) tunes ecdysteroidogenesis and mediates 20-hydroxyecdysone signaling during larval-pupal metamorphosis. Insect Biochem. Mol. Biol. 2018, 94, 50–60. [Google Scholar] [CrossRef]
- Chan, S.-M.; Gu, P.-L.; Chu, K.-H.; Tobe, S.S. Crustacean neuropeptide genes of the CHH/MIH/GIH family: Implications from molecular studies. Gen. Comp. Endocrinol. 2003, 134, 214–219. [Google Scholar] [CrossRef]
- Chang, E.S.; Mykles, D.L. Regulation of crustacean molting: A review and our perspectives. Gen. Comp. Endocrinol. 2011, 172, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Webster, S.G.; Keller, R.; Dircksen, H. The CHH-superfamily of multifunctional peptide hormones controlling crustacean metabolism, osmoregulation, moulting, and reproduction. Gen. Comp. Endocrinol. 2012, 175, 217–233. [Google Scholar] [CrossRef]
- Techa, S.; Chung, J.S. Ecdysteroids regulate the levels of Molt-Inhibiting Hormone (MIH) expression in the blue crab, Callinectes sapidus. PLoS ONE 2015, 10, e0117278. [Google Scholar] [CrossRef]
- Raviv, S.; Parnes, S.; Sagi, A. Coordination of Reproduction and Molt in Decapods; Mente, E., Ed.; Reproductive Biology of Crustaceans Sciences Publishers: Enfield, NH, USA, 2008; pp. 365–390. [Google Scholar]
- Laufer, H.; Wainwright, G.; Young, N.J.; Sagi, A.; Ahl, J.S.B.; Rees, H.H. Ecdysteroids and juvenoids in two male morphotypes of Libinia emarginata. Insect Biochem. Mol. Biol. 1993, 23, 171–174. [Google Scholar] [CrossRef]
- Parnes, S.; Raviv, S.; Shechter, A.; Sagi, A. Males also have their time of the month! Cyclic disposal of old spermatophores, timed by the molt cycle, in a marine shrimp. J. Exp. Biol. 2006, 209, 4974–4983. [Google Scholar] [CrossRef]
- Dominik, M.-C.; Stephanie, A.W.; Klaus, H.H. Ecdysteroid levels in Daphnia magna during a molt cycle: Determination by radioimmunoassay (RIA) and liquid chromatography–mass spectrometry (LC–MS). Gen. Comp. Endocrinol. 2007, 151, 66–71. [Google Scholar] [CrossRef]
- Teruaki, N.; Haruyuki, S. Regulation of ecdysteroid secretion from the Y-organ by molt-inhibiting hormone in the American crayfish, Procambarus clarkii. Gen. Comp. Endocrinol. 2004, 135, 358–364. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, X.; Wei, J.; Sun, X.; Yuan, J.; Xiang, J. Whole transcriptome analysis provides insights into molecular mechanisms for molting in Litopenaeus vannamei. PLoS ONE 2015, 10, e0144350. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.H.; Li, S.H.; Wang, Z.W.; Li, F.H.; Xiang, J.H. An eclosion hormone-like gene participates in the molting process of Palaemonid shrimp Exopalaemon carinicauda. Dev. Genes Evol. 2017, 227, 189–199. [Google Scholar] [CrossRef] [PubMed]


| Sample | Raw Reads | Clean Reads | Q20 (%) | Q30 (%) | Total Mapped | GC Content (%) |
|---|---|---|---|---|---|---|
| eAG1 | 39,144,350 | 38,623,336 | 97.64 | 93.61 | 32,983,645 (88.06%) | 48.65 |
| eAG2 | 38,000,328 | 37,384,452 | 97.48 | 93.28 | 31,424,401 (87.06%) | 47.65 |
| eAG3 | 37,794,382 | 37,206,858 | 97.53 | 93.47 | 32,130,046 (88.44%) | 48.77 |
| cAG1 | 42,634,574 | 41,927,468 | 97.50 | 93.34 | 36,357,115 (88.87%) | 49.85 |
| cAG2 | 38,989,158 | 38,455,262 | 97.74 | 93.84 | 31,867,585 (86.36%) | 49.13 |
| cAG3 | 44,795,738 | 43,610,558 | 97.53 | 93.63 | 37,288,555 (88.04%) | 50.29 |
| Categories | Gene Symbol | Accession Number | Annotation | Log2 (Fold Change ESA/Control) |
|---|---|---|---|---|
| Sexual development | Lv-IAG | MSTRG.32535 | insulin-like androgenic gland hormone | 2.41 |
| Lv-Mrr1 | ROT78373.1 | male reproductive-related protein A | 3.09 | |
| Lv-Mrr2 | MSTRG.11644 | male reproductive-related protein A | 3.3 | |
| Lv-GGCX | ROT82257.1 | Vitamin K-dependent gamma-carboxylase | 3.3 | |
| Lv-Calu | ROT83784.1 | Calumenin | 2.76 | |
| Lv-Hsp90-1 | ROT62469.1 | heat shock protein 90 | 2.76 | |
| Lv-Hsp70 | ROT68844.1 | heat shock protein 70 | 3.1 | |
| Lv-Hsp90-2 | ROT76137.1 | heat shock protein 90 | 2.63 | |
| Lv-ZSCAN31 | ROT76330.1 | zinc finger and SCAN domain-containing protein 31 | −4.18 | |
| Lv-ZCWPW1 | MSTRG.12712 | zinc finger/CW type with PWWP domain 1 | −1.89 | |
| Endocrine system | Lv-Hr4 | ROT82234.1 | hormone receptor 4 | −1.75 |
| Lv-SPI | ROT62953.1 | serine proteinase inhibitor | 2.47 | |
| Lv-SPI3 | ROT75925.1 | serine proteinase inhibitor-3 | −6.43 | |
| Lv-DBH1 | ROT77266.1 | dopamine beta-hydroxylase | 2.35 | |
| Lv-DBH2 | ROT77268.1 | dopamine beta-hydroxylase | 2.75 | |
| Lv-DBH3 | ROT77267.1 | dopamine beta-hydroxylase | 2.54 | |
| Lv-GRWD1 | ROT82204.1 | glutamate-rich WD repeat-containing protein 1 | 2.47 | |
| Lv-LRP2 | ROT73017.1 | lipoprotein receptor-related protein 2 | −4.25 | |
| Lv-PA2G4 | ROT64438.1 | proliferation-associated protein 2G4 | 1.79 | |
| Lv-ANK3 | ROT77030.1 | ankyrin-3-like isoform X1 | −3.05 |
| Gene Symbol | Primer Name | Nucleotide Sequences (5′-3′) | Annealing Temperature (°C) |
|---|---|---|---|
| Lv-IAG | Lv-IAG-qF | AGTGTCAAGGTCAGCCGATAC | 54.5 °C |
| Lv-IAG-qR | CGAGATTCCACGTTGGATTCAG | ||
| Lv-18S | 18S-F | TATACGCTAGTGGAGCTGGAA | 54.9 °C |
| 18S-R | GGGGAGGTAGTGACGAAAAAT | ||
| Lv-Hr4 | Lv-Hr4-F | AGACTCCCCAGCAGCTATGA | 55.0 °C |
| Lv-Hr4-R | GTTGCGCTGTGCCTTTGTAA | ||
| Lv-DBH1 | Lv-DBH1-qF | TCGAAGCCGAAGACAGAACC | 56.0 °C |
| Lv-DBH1-qR | AGGCGCTCACTTCTCTCAAC | ||
| Lv-DBH2 | Lv-DBH2-qF | CAGCACCGATTTCAGCGATG | 55.0 °C |
| Lv-DBH2-qR | ATCCTCTTCCCCTGGAGTCC | ||
| Lv-DBH3 | Lv-DBH3-qF | TATGGTGCCAACATGCCCAT | 54.0 °C |
| Lv-DBH3-qR | GGGACGATTTGGATTGGGGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, K.; Li, S.; Xiang, J.; Sagi, A.; Li, F. Transcriptome Analysis Reveals the Endocrine Regulation on the Expression of IAG in Litopenaeus vannamei. J. Mar. Sci. Eng. 2021, 9, 677. https://doi.org/10.3390/jmse9060677
Chen K, Li S, Xiang J, Sagi A, Li F. Transcriptome Analysis Reveals the Endocrine Regulation on the Expression of IAG in Litopenaeus vannamei. Journal of Marine Science and Engineering. 2021; 9(6):677. https://doi.org/10.3390/jmse9060677
Chicago/Turabian StyleChen, Kangxuan, Shihao Li, Jianhai Xiang, Amir Sagi, and Fuhua Li. 2021. "Transcriptome Analysis Reveals the Endocrine Regulation on the Expression of IAG in Litopenaeus vannamei" Journal of Marine Science and Engineering 9, no. 6: 677. https://doi.org/10.3390/jmse9060677






