Sialylation of Human Natural Killer (NK) Cells Is Regulated by IL-2
Abstract
:1. Introduction
2. Experimental Section
2.1. Cells and Cell Culture
2.2. PCR and Quantitative Real-Time PCR Analysis
2.3. Flow Cytometry Analysis
| Gene | Product | Primer |
|---|---|---|
| ST8SIA1 | 334 bp | forward: AATCCCAGCATAATTCGGCAAAG |
| reverse: AGAAGGGCCAGAAGCCATAG | ||
| ST8SIA2 | 340 bp | forward: TCTTCGATCGAGACAGCACCA |
| reverse: CACAGGATGCTGCCATTGAGG | ||
| ST8SIA3 | 323 bp | forward: ATTTGGCGCTTTCCGTTTGG |
| reverse: GCAACATGTCAACAGGTACTGG | ||
| ST8SIA4 | 303 bp | forward: TCTAGCTCCTGTGGTGGAGTT |
| reverse: TTGGTCAGCCAGTAACCTCTG | ||
| ST8SIA5 | 328 bp | forward: AGTCTACTCTGTCCAGGTGCT |
| reverse: ACAGTGACCACATCCGTCTTC | ||
| ST8SIA6 | 342 bp | forward: GTAACCTACCCCCAACCACAG |
| reverse: TCATCAAGCCGGTGGACAAG | ||
| ST6GALNAC1 | 356 bp | forward: AGCCTCGGTGGGATTTTGAG |
| reverse: GGAGTTGTTCAGGATGCCCC | ||
| ST6GALNAC2 | 303 bp | forward: TTTGCCCTGTACTTCTCGGC |
| reverse: GGAGGCGATGACTTGGTGAG | ||
| ST6GALNAC3 | 339 bp | forward: TGGCCTGCATCCTGAAGAGAA |
| reverse: CTTTGGTGGGGGCATTGTTC | ||
| ST6GALNAC4 | 299 bp | forward: CGTGGTCTATGGGATGGTCAG |
| reverse: TGGAGTGTGATGGCTTGGGA | ||
| ST6GALNAC5 | 302 bp | forward: AGGGCACCGTGTTCATCTTC |
| reverse: GTGATTGGGATCCCTGCAGAA | ||
| ST6GALNAC6 | 339 bp | forward: CGGTCAGCAGTGTTCGTGA |
| reverse: GCGGTAGGTGGTCTTGTTGC | ||
| ST6GAL1 | 332 bp | forward: TCCCAAAGTGGTACCAGAATCC |
| reverse: CTTCTCATAGAGCAGCGGGT | ||
| ST6GAL2 | 336 bp | forward: TCTGCTCCTACACGTGGTTATG |
| reverse: AGAAGATGGTGGGTTTGGTTGA | ||
| ST3GAL1 | 321 bp | forward: ATGTTGGGACCAAGACCACC |
| reverse: ACAAGTCCACCTCATCGCAG | ||
| ST3GAL2 | 333 bp | forward: CAGATAGTGCCTGGCGAGAA |
| reverse: CACTGGGGCGTAGGTGAATC | ||
| ST3GAL3 | 337 bp | forward: CCTTTCGCAAGTGGGCTAGA |
| reverse: AGAGAATCGCGCTCGTACTG | ||
| ST3GAL4 | 335 bp | forward: CCTACAACAAGAAGCAGACCATTC |
| reverse: CTGGATCTCGGCTCCATAAGAG | ||
| ST3GAL5 | 333 bp | forward: AACAGTGCACCAGTTGAGGG |
| reverse: GCCCCAGAACCTTGACTGAG | ||
| ST3GAL6 | 297 bp | forward: GACCTCAAGAGTCCTTTGCAC |
| reverse: TTCACAGAAATTAAGCTGGTGGTT | ||
| IFN-γ | 330 bp | forward: GAATTGGAAAGAGGAGAGTGACA |
| reverse: TACTGGGATGCTCTTCGACCT | ||
| B2M1 | 86 bp | forward: TGCTGTCTCCATGTTTGATGTATCT |
| reverse: TCTCTGCTCCCCACCTCTAAGT |
2.4. Western Blot Analysis
2.5. Statistical Analysis.
3. Results
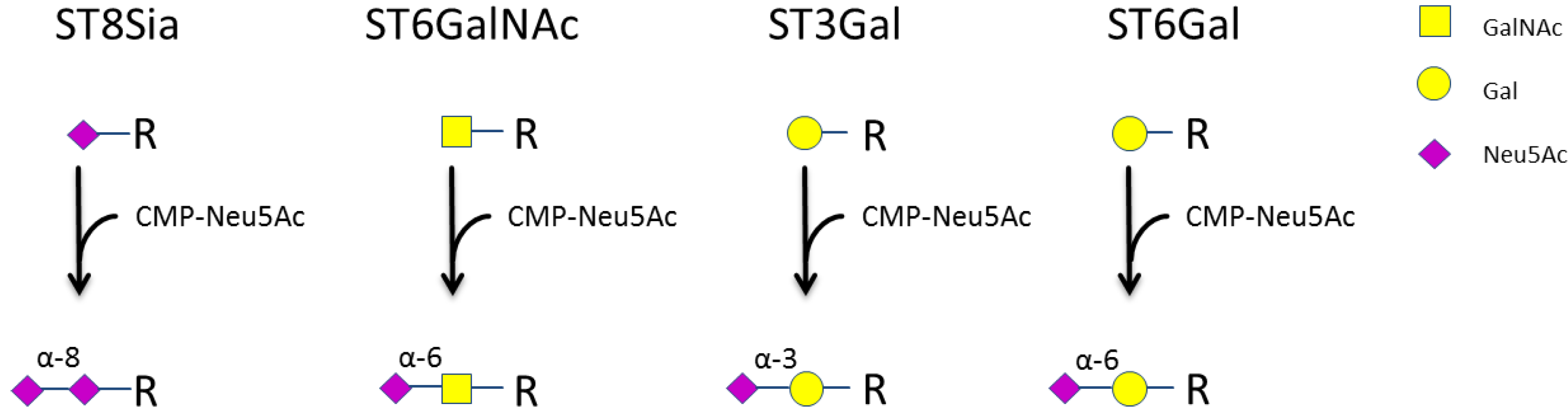
3.1. Expression of Sialyltransferases in Human NK Cells
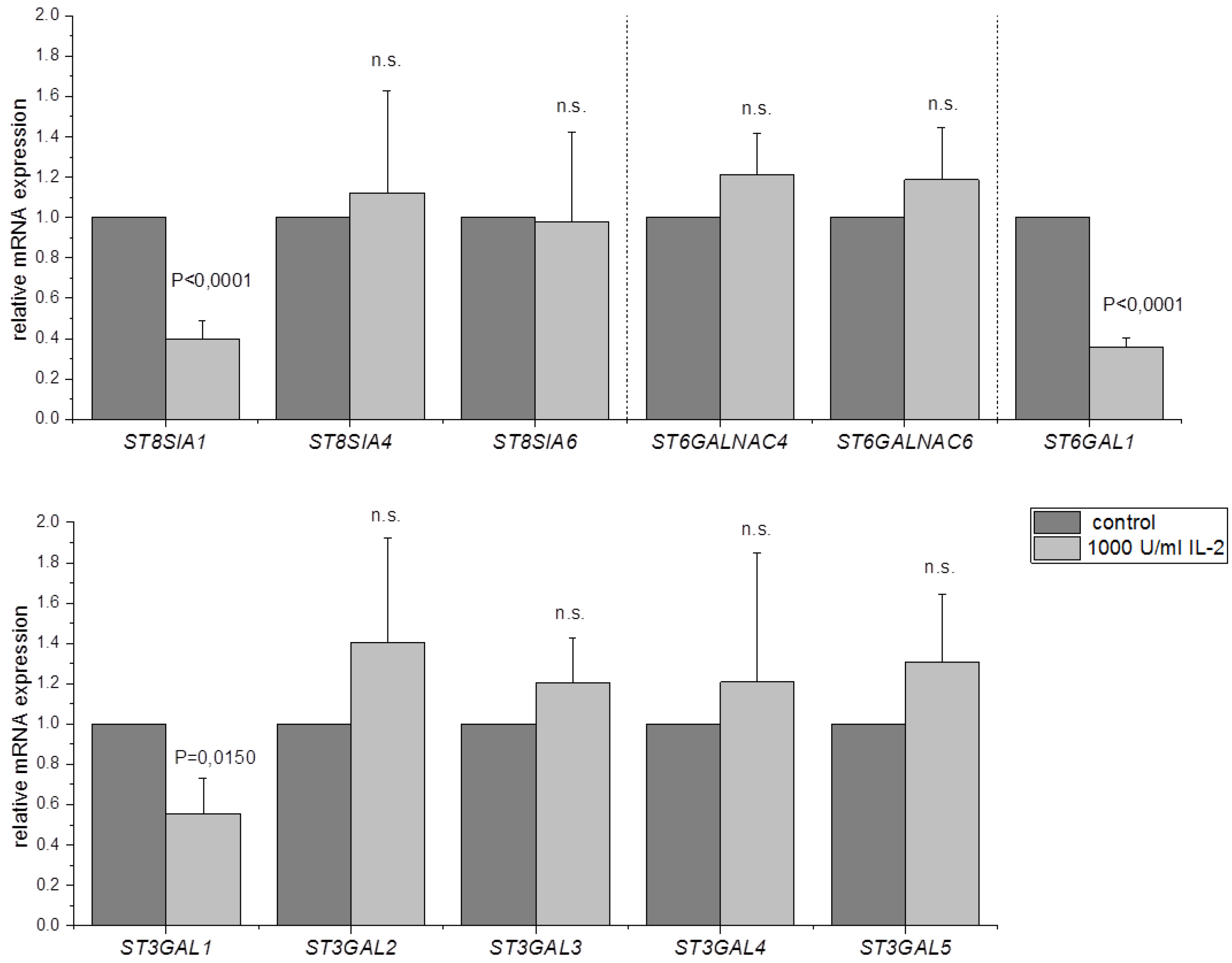
3.2. ST8SIA1, ST6GAL1 and ST3GAL1 Are Downregulated after IL-2 Stimulation
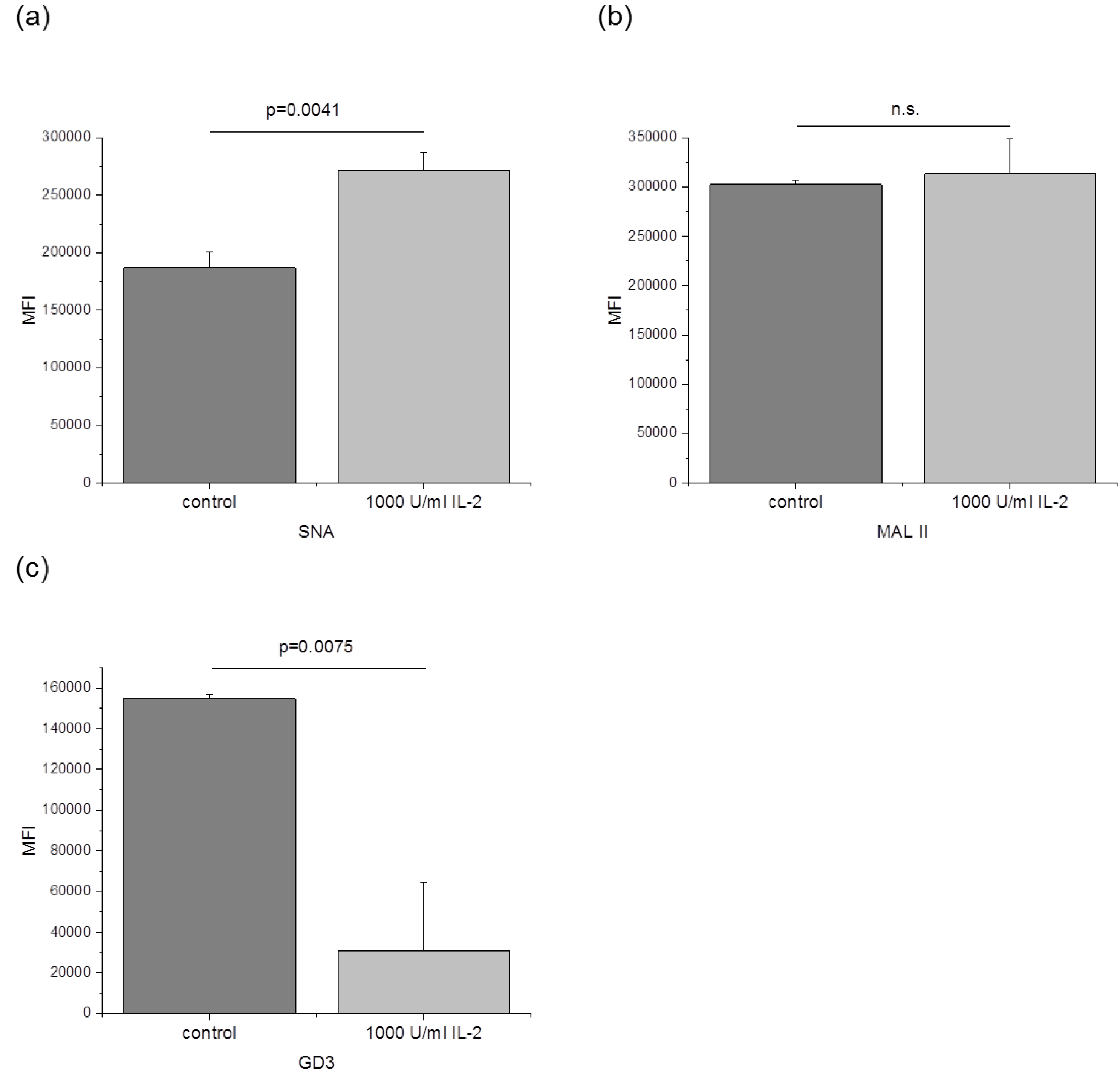
3.3. The Amount of α-2,6 Linked Sialic Acids and GD3 Is Changed after IL-2 Stimulation
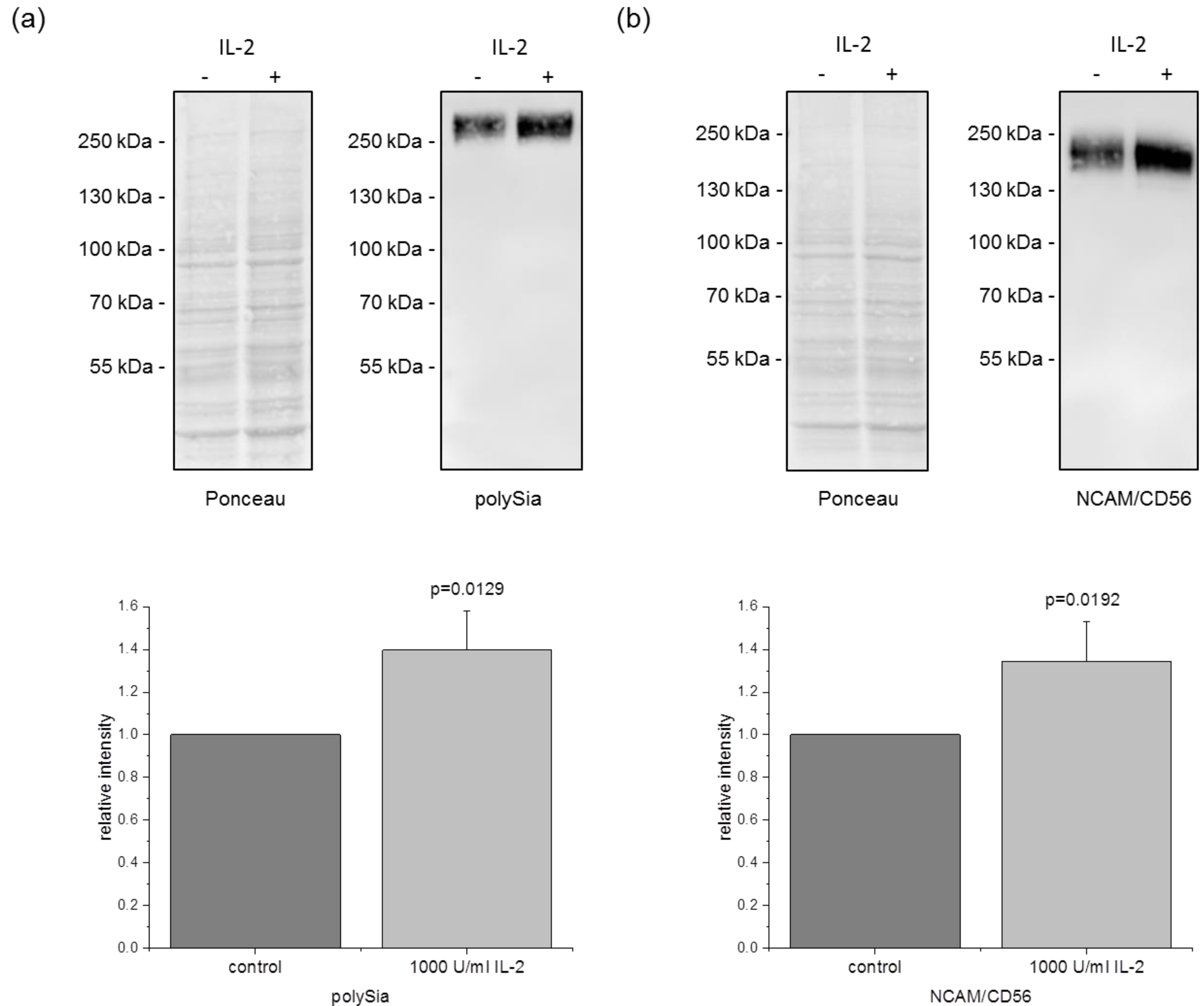
3.4. PolySia and NCAM/CD56 Expression Is Increased after IL-2 Stimulation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rudd, P.M.; Elliott, T.; Cresswell, P.; Wilson, I.A.; Dwek, R.A. Glycosylation and the immune system. Science 2001, 291, 2370–2376. [Google Scholar] [CrossRef]
- Marth, J.D.; Grewal, P.K. Mammalian glycosylation in immunity. Nat. Rev. Immunol. 2008, 8, 874–887. [Google Scholar] [CrossRef] [Green Version]
- Varki, A.; Schauer, R. Sialic Acids. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor: New York, NY, USA, 2009; ISBN 9780879697709. [Google Scholar]
- Wang, B.; Brand-Miller, J. The role and potential of sialic acid in human nutrition. Eur. J. Clin. Nutr. 2003, 57, 1351–1369. [Google Scholar] [CrossRef] [Green Version]
- Hinderlich, S.; Weidemann, W.; Yardeni, T.; Horstkorte, R.; Huizing, M. UDP-GlcNAc 2-Epimerase/ManNAc Kinase (GNE): A Master Regulator of Sialic Acid Synthesis. Top. Curr. Chem. 2015, 366, 97–137. [Google Scholar] [CrossRef] [Green Version]
- Nji, E.; Gulati, A.; Qureshi, A.A.; Coincon, M.; Drew, D. Structural basis for the delivery of activated sialic acid into Golgi for sialyation. Nat. Struct. Mol. Biol. 2019, 26, 415–423. [Google Scholar] [CrossRef]
- Harduin-Lepers, A.; Krzewinski-Recchi, M.-A.; Colomb, F.; Foulquier, F.; Groux-Degroote, S.; Delannoy, P. Sialyltransferases functions in cancers. Front. Biosci. (Elite Ed.) 2012, 4, 499–515. [Google Scholar] [CrossRef]
- Takashima, S.; Tsuji, S.; Tsujimoto, M. Characterization of the second type of human beta-galactoside alpha 2,6-sialyltransferase (ST6Gal II), which sialylates Galbeta 1,4GlcNAc structures on oligosaccharides preferentially. Genomic analysis of human sialyltransferase genes. J. Biol. Chem. 2002, 277, 45719–45728. [Google Scholar] [CrossRef] [Green Version]
- Harduin-Lepers, A. Comprehensive Analysis of Sialyltransferases in Vertebrate Genomes. Glycobiol. Insights 2010, 2, 29–61. [Google Scholar] [CrossRef]
- Varki, A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature 2007, 446, 1023–1029. [Google Scholar] [CrossRef]
- Lübbers, J.; Rodríguez, E.; van Kooyk, Y. Modulation of Immune Tolerance via Siglec-Sialic Acid Interactions. Front. Immunol. 2018, 9, 2807. [Google Scholar] [CrossRef] [Green Version]
- Farag, S.S.; Caligiuri, M.A. Human natural killer cell development and biology. Blood Rev. 2006, 20, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Pegram, H.J.; Andrews, D.M.; Smyth, M.J.; Darcy, P.K.; Kershaw, M.H. Activating and inhibitory receptors of natural killer cells. Immunol. Cell Biol. 2011, 89, 216–224. [Google Scholar] [CrossRef]
- Cooper, M.A.; Fehniger, T.A.; Caligiuri, M.A. The biology of human natural killer-cell subsets. Trends Immunol. 2001, 22, 633–640. [Google Scholar] [CrossRef]
- Kerdiles, Y.; Ugolini, S.; Vivier, E. T cell regulation of natural killer cells. J. Exp. Med. 2013, 210, 1065–1068. [Google Scholar] [CrossRef] [PubMed]
- Krishnaraj, R.; Bhooma, T. Cytokine sensitivity of human NK cells during immunosenescence. 2. IL2-induced Interferon gamma secretion. Immunol. Lett. 1996, 50, 59–63. [Google Scholar] [CrossRef]
- Lehmann, C.; Zeis, M.; Uharek, L. Activation of natural killer cells with interleukin 2 (IL-2) and IL-12 increases perforin binding and subsequent lysis of tumour cells. Br. J. Haematol. 2001, 114, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Huenecke, S.; Zimmermann, S.Y.; Kloess, S.; Esser, R.; Brinkmann, A.; Tramsen, L.; Koenig, M.; Erben, S.; Seidl, C.; Tonn, T.; et al. IL-2-driven regulation of NK cell receptors with regard to the distribution of CD16+ and CD16- subpopulations and in vivo influence after haploidentical NK cell infusion. J. Immunother. 2010, 33, 200–210. [Google Scholar] [CrossRef]
- Hromadnikova, I.; Pirkova, P.; Sedlackova, L. Influence of In Vitro IL-2 or IL-15 Alone or in Combination with Hsp-70-Derived 14-mer Peptide (TKD) on the Expression of NK Cell Activatory and Inhibitory Receptors. Mediat. Inflamm. 2013, 2013. [Google Scholar] [CrossRef] [Green Version]
- de Rham, C.; Ferrari-Lacraz, S.; Jendly, S.; Schneiter, G.; Dayer, J.-M.; Villard, J. The proinflammatory cytokines IL-2, IL-15 and IL-21 modulate the repertoire of mature human natural killer cell receptors. Arthritis Res. Ther. 2007, 9, R125. [Google Scholar] [CrossRef] [Green Version]
- Rosenstock, P.; Horstkorte, R.; Gnanapragassam, V.S.; Harth, J.; Kielstein, H. Siglec-7 expression is reduced on a natural killer (NK) cell subset of obese humans. Immunol. Res. 2017, 4, 579. [Google Scholar] [CrossRef] [Green Version]
- Moebius, J.M.; Widera, D.; Schmitz, J.; Kaltschmidt, C.; Piechaczek, C. Impact of polysialylated CD56 on natural killer cell cytotoxicity. BMC Immunol. 2007, 8, 13. [Google Scholar] [CrossRef] [Green Version]
- Jandus, C.; Boligan, K.F.; Chijioke, O.; Liu, H.; Dahlhaus, M.; Démoulins, T.; Schneider, C.; Wehrli, M.; Hunger, R.E.; Baerlocher, G.M.; et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J. Clin. Investig. 2014, 124, 1810–1820. [Google Scholar] [CrossRef] [Green Version]
- Razi, N.; Varki, A. Masking and unmasking of the sialic acid-binding lectin activity of CD22 (Siglec-2) on B lymphocytes. Proc. Natl. Acad. Sci. USA 1998, 95, 7469–7474. [Google Scholar] [CrossRef] [Green Version]
- Razi, N.; Varki, A. Cryptic sialic acid binding lectins on human blood leukocytes can be unmasked by sialidase treatment or cellular activation. Glycobiology 1999, 9, 1225–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [Green Version]
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; van Roy, N.; Paepe, A.D.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Matsuo, Y.; Drexler, H.G. Immunoprofiling of cell lines derived from natural killer-cell and natural killer-like T-cell leukemia–lymphoma. Leuk. Res. 2003, 27, 935–945. [Google Scholar] [CrossRef]
- Dybkaer, K.; Iqbal, J.; Zhou, G.; Geng, H.; Xiao, L.; Schmitz, A.; d’Amore, F.; Chan, W.C. Genome wide transcriptional analysis of resting and IL2 activated human natural killer cells: Gene expression signatures indicative of novel molecular signaling pathways. BMC Genom. 2007, 8, 230. [Google Scholar] [CrossRef] [Green Version]
- Kaszubowska, L.; Wierzbicki, P.M.; Karsznia, S.; Damska, M.; Ślebioda, T.J.; Foerster, J.; Kmieć, Z. Optimal reference genes for qPCR in resting and activated human NK cells—Flow cytometric data correspond to qPCR gene expression analysis. J. Immunol. Methods 2015, 422, 125–129. [Google Scholar] [CrossRef]
- Hodge, D.L.; Schill, W.B.; Wang, J.M.; Blanca, I.; Reynolds, D.A.; Ortaldo, J.R.; Young, H.A. IL-2 and IL-12 Alter NK Cell Responsiveness to IFN- -Inducible Protein 10 by Down-Regulating CXCR3 Expression. J. Immunol. 2002, 168, 6090–6098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avril, T.; North, S.J.; Haslam, S.M.; Willison, H.J.; Crocker, P.R. Probing the cis interactions of the inhibitory receptor Siglec-7 with alpha2,8-disialylated ligands on natural killer cells and other leukocytes using glycan-specific antibodies and by analysis of alpha2,8-sialyltransferase gene expression. J. Leukoc. Biol. 2006, 80, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Comelli, E.M.; Sutton-Smith, M.; Yan, Q.; Amado, M.; Panico, M.; Gilmartin, T.; Whisenant, T.; Lanigan, C.M.; Head, S.R.; Goldberg, D.; et al. Activation of murine CD4+ and CD8+ T lymphocytes leads to dramatic remodeling of N-linked glycans. J. Immunol. 2006, 177, 2431–2440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyagi, T.; Yamaguchi, K. Mammalian sialidases: Physiological and pathological roles in cellular functions. Glycobiology 2012, 22, 880–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Zhang, S.; Zou, X.; Lu, J.; Yang, X.; Xu, Z.; Shan, A.; Jia, W.; Liu, F.; Yan, X.; et al. The β-galactoside α2,6-sialyltranferase 1 (ST6GAL1) inhibits the colorectal cancer metastasis by stabilizing intercellular adhesion molecule-1 via sialylation. Cancer Manag. Res. 2019, 11, 6185–6199. [Google Scholar] [CrossRef] [Green Version]
- Margraf-Schönfeld, S.; Böhm, C.; Watzl, C. Glycosylation affects ligand binding and function of the activating natural killer cell receptor 2B4 (CD244) protein. J. Biol. Chem. 2011, 286, 24142–24149. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, K.; Kurata, K.; Kojima, N.; Kurosawa, N.; Ohta, S.; Hanai, N.; Tsuji, S.; Nishi, T. Expression cloning of a GM3-specific alpha-2,8-sialyltransferase (GD3 synthase). J. Biol. Chem. 1994, 269, 15950–15956. [Google Scholar]
- Liu, J.; Zheng, X.; Pang, X.; Li, L.; Wang, J.; Yang, C.; Du, G. Ganglioside GD3 synthase (GD3S), a novel cancer drug target. Acta Pharm. Sin. B 2018, 8, 713–720. [Google Scholar] [CrossRef]
- Zhang, T.; de Waard, A.A.; Wuhrer, M.; Spaapen, R.M. The Role of Glycosphingolipids in Immune Cell Functions. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Nicoll, G.; Avril, T.; Lock, K.; Furukawa, K.; Bovin, N.; Crocker, P.R. Ganglioside GD3 expression on target cells can modulate NK cell cytotoxicity via siglec-7-dependent and -independent mechanisms. Eur. J. Immunol. 2003, 33, 1642–1648. [Google Scholar] [CrossRef]
- Drake, P.M.; Nathan, J.K.; Stock, C.M.; Chang, P.V.; Muench, M.O.; Nakata, D.; Reader, J.R.; Gip, P.; Golden, K.P.K.; Weinhold, B.; et al. Polysialic acid, a glycan with highly restricted expression, is found on human and murine leukocytes and modulates immune responses. J. Immunol. 2008, 181, 6850–6858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziegler, S.; Weiss, E.; Schmitt, A.-L.; Schlegel, J.; Burgert, A.; Terpitz, U.; Sauer, M.; Moretta, L.; Sivori, S.; Leonhardt, I.; et al. CD56 Is a Pathogen Recognition Receptor on Human Natural Killer Cells. Sci. Rep. 2017, 7, 6138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mace, E.M.; Gunesch, J.T.; Dixon, A.; Orange, J.S. Human NK cell development requires CD56-mediated motility and formation of the developmental synapse. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| NK-92 (n = 3) | NKL (n = 3) | KHYG-1 (n = 3) | Primary NK Cells (From 3 Donors) | |
|---|---|---|---|---|
| ST8SIA1 | + | + | + | 3/3 |
| ST8SIA2 | - | - | - | - |
| ST8SIA3 | - | - | - | - |
| ST8SIA4 | + | + | + | 3/3 |
| ST8SIA5 | - | - | - | - |
| ST8SIA6 | + | + | + | 3/3 |
| ST6GALNAC1 | - | - | - | - |
| ST6GALNAC2 | - | - | - | - |
| ST6GALNAC3 | - | - | - | - |
| ST6GALNAC4 | + | + | + | 3/3 |
| ST6GALNAC5 | - | - | - | - |
| ST6GALNAC6 | + | + | + | 3/3 |
| ST6GAL1 | + | + | + | 3/3 |
| ST6GAL2 | - | - | - | 2/3 |
| ST3GAL1 | + | + | + | 3/3 |
| ST3GAL2 | + | + | + | 3/3 |
| ST3GAL3 | + | + | + | 3/3 |
| ST3GAL4 | + | + | + | 3/3 |
| ST3GAL5 | + | + | + | 3/3 |
| ST3GAL6 | - | - | - | 2/3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosenstock, P.; Bork, K.; Massa, C.; Selke, P.; Seliger, B.; Horstkorte, R. Sialylation of Human Natural Killer (NK) Cells Is Regulated by IL-2. J. Clin. Med. 2020, 9, 1816. https://doi.org/10.3390/jcm9061816
Rosenstock P, Bork K, Massa C, Selke P, Seliger B, Horstkorte R. Sialylation of Human Natural Killer (NK) Cells Is Regulated by IL-2. Journal of Clinical Medicine. 2020; 9(6):1816. https://doi.org/10.3390/jcm9061816
Chicago/Turabian StyleRosenstock, Philip, Kaya Bork, Chiara Massa, Philipp Selke, Barbara Seliger, and Rüdiger Horstkorte. 2020. "Sialylation of Human Natural Killer (NK) Cells Is Regulated by IL-2" Journal of Clinical Medicine 9, no. 6: 1816. https://doi.org/10.3390/jcm9061816




