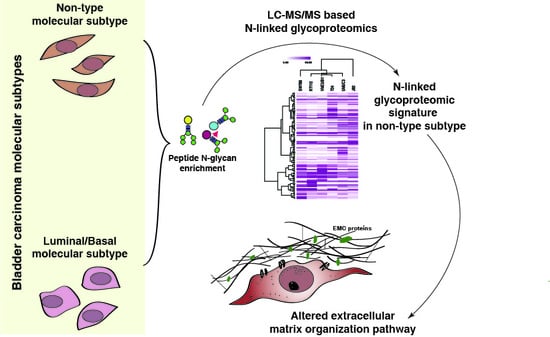
N-Glycoproteomic Profiling Reveals Alteration In Extracellular Matrix Organization In Non-Type Bladder Carcinoma
Abstract
:1. Introduction
2. Experimental Section
2.1. Cell Culture
2.2. Cell Lysis and Protein Extraction
2.3. In-Solution Trypsin Digestion of Proteins and TMT Labeling
2.4. N-Glycan Enrichment
2.5. LC-MS/MS Analysis
2.6. Data Analysis
2.7. Statistical Analysis
2.8. Clustering the Molecular Subtypes
2.9. Protein-Protein Interaction Network Analysis
2.10. Reactome Pathway Analysis
3. Results
3.1. Quantitative Analysis of N-Glycoprotein in Bladder Carcinoma Cell Lines
3.2. N-Linked Glycosylation in Aggressive Non-Type Bladder Carcinoma
3.3. Unique N-Linked Glycosylation Signature in Aggressive Non-Type Bladder Carcinoma Cell Lines
3.4. Interaction Network Clusteres in the Non-Type Bladder Carcinoma Cell Lines
3.5. Reactome Pathway Analysis Identified Dysregulation of Extracellular Matrix Organization Pathway Enriched in the Non-Type Bladder Carcinoma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sharma, J.; Deb, B.; George, I.A.; Kapil, S.; Coral, K.; Kakkar, N.; Pattanaik, S.; Mandal, A.K.; Mavuduru, R.S.; Kumar, P. Somatic Mutations Profile of a Young Patient with Metastatic Urothelial Carcinoma Reveals Mutations in Genes Involved in Ion Channels. Front. Oncol. 2019, 9, 435. [Google Scholar] [CrossRef] [PubMed]
- Noyes, K.; Singer, E.A.; Messing, E.M. Healthcare economics of bladder cancer: Cost-enhancing and cost-reducing factors. Curr. Opin. Urol. 2008, 18, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Sievert, K.D.; Amend, B.; Nagele, U.; Schilling, D.; Bedke, J.; Horstmann, M.; Hennenlotter, J.; Kruck, S.; Stenzl, A. Economic aspects of bladder cancer: What are the benefits and costs? World J. Urol. 2009, 27, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nandi, S.; Tan, T.Z.; Ler, S.G.; Chia, K.S.; Lim, W.Y.; Butow, Z.; Vordos, D.; De la Taille, A.; Al-Haddawi, M.; et al. Highly sensitive and specific novel biomarkers for the diagnosis of transitional bladder carcinoma. Oncotarget 2015, 6, 13539–13549. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.Z.; Rouanne, M.; Tan, K.T.; Huang, R.Y.; Thiery, J.P. Molecular Subtypes of Urothelial Bladder Cancer: Results from a Meta-cohort Analysis of 2411 Tumors. Eur. Urol. 2019, 75, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Kolbl, A.C.; Andergassen, U.; Jeschke, U. The Role of Glycosylation in Breast Cancer Metastasis and Cancer Control. Front. Oncol. 2015, 5, 219. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, P.K.; Bouchie, M.P.; Kukuruzinska, M.A. N-glycosylation gene DPAGT1 is a target of the Wnt/beta-catenin signaling pathway. J. Biol. Chem. 2010, 285, 31164–31173. [Google Scholar] [CrossRef]
- Huang, S.; Wang, D.; Zhang, S.; Huang, X.; Wang, D.; Ijaz, M.; Shi, Y. Tunicamycin potentiates paclitaxel-induced apoptosis through inhibition of PI3K/AKT and MAPK pathways in breast cancer. Cancer Chemother. Pharmacol. 2017, 80, 685–696. [Google Scholar] [CrossRef]
- Zhao, Y.; Sato, Y.; Isaji, T.; Fukuda, T.; Matsumoto, A.; Miyoshi, E.; Gu, J.; Taniguchi, N. Branched N-glycans regulate the biological functions of integrins and cadherins. FEBS J. 2008, 275, 1939–1948. [Google Scholar] [CrossRef]
- Pinho, S.S.; Figueiredo, J.; Cabral, J.; Carvalho, S.; Dourado, J.; Magalhaes, A.; Gartner, F.; Mendonfa, A.M.; Isaji, T.; Gu, J.; et al. E-cadherin and adherens-junctions stability in gastric carcinoma: Functional implications of glycosyltransferases involving N-glycan branching biosynthesis, N-acetylglucosaminyltransferases III and V. Biochim. Biophys. Acta 2013, 1830, 2690–2700. [Google Scholar] [CrossRef]
- Takeuchi, H.; Haltiwanger, R.S. Significance of glycosylation in Notch signaling. Biochem. Biophys. Res. Commun. 2014, 453, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dennis, J.W.; Nabi, I.R.; Demetriou, M. Metabolism, cell surface organization, and disease. Cell 2009, 139, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, R.; Peixoto, A.; Gaiteiro, C.; Fernandes, E.; Neves, M.; Lima, L.; Santos, L.L.; Ferreira, J.A. Over forty years of bladder cancer glycobiology: Where do glycans stand facing precision oncology? Oncotarget 2017, 8, 91734–91764. [Google Scholar] [CrossRef] [PubMed]
- Kruger, S.; Weitsch, G.; Buttner, H.; Matthiensen, A.; Bohmer, T.; Marquardt, T.; Sayk, F.; Feller, A.C.; Bohle, A. HER2 overexpression in muscle-invasive urothelial carcinoma of the bladder: Prognostic implications. Int. J. Cancer 2002, 102, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Sanguedolce, F.; Russo, D.; Mancini, V.; Selvaggio, O.; Calo, B.; Carrieri, G.; Cormio, L. Human Epidermal Growth Factor Receptor 2 in Non-Muscle Invasive Bladder Cancer: Issues in Assessment Methods and Its Role as Prognostic/Predictive Marker and Putative Therapeutic Target: A Comprehensive Review. Urol. Int. 2019, 102, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Schwaederle, M.; Arguello, D.; Millis, S.Z.; Gatalica, Z.; Kurzrock, R. HER2 expression status in diverse cancers: Review of results from 37,992 patients. Cancer Metastasis Rev. 2015, 34, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Yoneyama, T.; Noro, D.; Imanishi, K.; Kojima, Y.; Hatakeyama, S.; Tobisawa, Y.; Mori, K.; Yamamoto, H.; Imai, A.; et al. Aberrant N-Glycosylation Profile of Serum Immunoglobulins is a Diagnostic Biomarker of Urothelial Carcinomas. Int. J. Mol. Sci. 2017, 18, 2632. [Google Scholar] [CrossRef]
- Freire-de-Lima, L. Sweet and sour: The impact of differential glycosylation in cancer cells undergoing epithelial-mesenchymal transition. Front. Oncol. 2014, 4, 59. [Google Scholar] [CrossRef]
- Ding, Y.; Gelfenbeyn, K.; Freire-de-Lima, L.; Handa, K.; Hakomori, S.I. Induction of epithelial-mesenchymal transition with O-glycosylated oncofetal fibronectin. FEBS Lett. 2012, 586, 1813–1820. [Google Scholar] [CrossRef] [Green Version]
- Singh, C.; Shyanti, R.K.; Singh, V.; Kale, R.K.; Mishra, J.P.N.; Singh, R.P. Integrin expression and glycosylation patterns regulate cell-matrix adhesion and alter with breast cancer progression. Biochem. Biophys. Res. Commun. 2018, 499, 374–380. [Google Scholar] [CrossRef]
- Lemjabbar-Alaoui, H.; McKinney, A.; Yang, Y.W.; Tran, V.M.; Phillips, J.J. Glycosylation alterations in lung and brain cancer. Adv. Cancer Res. 2015, 126, 305–344. [Google Scholar] [CrossRef] [PubMed]
- Casey, R.C.; Oegema, T.R., Jr.; Skubitz, K.M.; Pambuccian, S.E.; Grindle, S.M.; Skubitz, A.P. Cell membrane glycosylation mediates the adhesion, migration, and invasion of ovarian carcinoma cells. Clin. Exp. Metastasis 2003, 20, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.L.; Hsu, W.M.; Huang, M.C.; Kadomatsu, K.; Nakagawara, A. Protein glycosylation in cancers and its potential therapeutic applications in neuroblastoma. J. Hematol. Oncol. 2016, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Esser, A.K.; Miller, M.R.; Huang, Q.; Meier, M.M.; Beltran-Valero de Bernabe, D.; Stipp, C.S.; Campbell, K.P.; Lynch, C.F.; Smith, B.J.; Cohen, M.B.; et al. Loss of LARGE2 disrupts functional glycosylation of alpha-dystroglycan in prostate cancer. J. Biol. Chem. 2013, 288, 2132–2142. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, C. Glycosylation in bladder cancer. Int. J. Clin. Oncol. 2008, 13, 308–313. [Google Scholar] [CrossRef]
- Guo, J.M.; Zhang, X.Y.; Chen, H.L.; Wang, G.M.; Zhang, Y.K. Structural alterations of sugar chains in urine fibronectin from bladder cancer patients and its enzymatic mechanism. J. Cancer Res. Clin. Oncol. 2001, 127, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Ishimura, H.; Takahashi, T.; Nakagawa, H.; Nishimura, S.; Arai, Y.; Horikawa, Y.; Habuchi, T.; Miyoshi, E.; Kyan, A.; Hagisawa, S.; et al. N-acetylglucosaminyltransferase V and beta1-6 branching N-linked oligosaccharides are associated with good prognosis of patients with bladder cancer. Clin. Cancer Res. 2006, 12, 2506–2511. [Google Scholar] [CrossRef]
- Warrick, J.I.; Walter, V.; Yamashita, H.; Chung, E.; Shuman, L.; Amponsa, V.O.; Zheng, Z.; Chan, W.; Whitcomb, T.L.; Yue, F.; et al. FOXA1, GATA3 and PPAR Cooperate to Drive Luminal Subtype in Bladder Cancer: A Molecular Analysis of Established Human Cell Lines. Sci. Rep. 2016, 6, 38531. [Google Scholar] [CrossRef]
- Deb, B.; Puttamallesh, V.N.; Gondkar, K.; Thiery, J.P.; Gowda, H.; Kumar, P. Phosphoproteomic Profiling Identifies Aberrant Activation of Integrin Signaling in Aggressive Non-Type Bladder Carcinoma. J. Clin. Med. 2019, 8, 703. [Google Scholar] [CrossRef]
- Tian, Y.; Zhou, Y.; Elliott, S.; Aebersold, R.; Zhang, H. Solid-phase extraction of N-linked glycopeptides. Nat. Protoc. 2007, 2, 334–339. [Google Scholar] [CrossRef] [Green Version]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef] [PubMed]
- Gu, J. Regulation of integrin functions by N-glycans. Yakugaku Zasshi 2007, 127, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Isaji, T.; Sato, Y.; Kariya, Y.; Fukuda, T. Importance of N-glycosylation on alpha5beta1 integrin for its biological functions. Biol. Pharm. Bull. 2009, 32, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Bane, S.M.; Kalraiya, R.D. Glycosylation of the laminin receptor (alpha3beta1) regulates its association with tetraspanin CD151: Impact on cell spreading, motility, degradation and invasion of basement membrane by tumor cells. Exp. Cell Res. 2014, 322, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yang, Z.; Du, Y.; Tang, H.; Chen, J.; Hu, D.; Fan, Z. BCMab1, a monoclonal antibody against aberrantly glycosylated integrin alpha3beta1, has potent antitumor activity of bladder cancer in vivo. Clin. Cancer Res. 2014, 20, 4001–4013. [Google Scholar] [CrossRef] [PubMed]
- Van der Horst, G.; van den Hoogen, C.; Buijs, J.T.; Cheung, H.; Bloys, H.; Pelger, R.C.; Lorenzon, G.; Heckmann, B.; Feyen, J.; Pujuguet, P.; et al. Targeting of alpha(v)-integrins in stem/progenitor cells and supportive microenvironment impairs bone metastasis in human prostate cancer. Neoplasia 2011, 13, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Manning, C.D.; Millar, H.; McCabe, F.L.; Ferrante, C.; Sharp, C.; Shahied-Arruda, L.; Doshi, P.; Nakada, M.T.; Anderson, G.M. CNTO 95, a fully human anti alphav integrin antibody, inhibits cell signaling, migration, invasion, and spontaneous metastasis of human breast cancer cells. Clin. Exp. Metastasis 2008, 25, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Vallo, S.; Rutz, J.; Kautsch, M.; Winkelmann, R.; Michaelis, M.; Wezel, F.; Bartsch, G.; Haferkamp, A.; Rothweiler, F.; Blaheta, R.A.; et al. Blocking integrin beta1 decreases adhesion in chemoresistant urothelial cancer cell lines. Oncol. Lett. 2017, 14, 5513–5518. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.B.; Chen, F.; Sima, N. Focal adhesion kinases crucially regulate TGFbeta-induced migration and invasion of bladder cancer cells via Src kinase and E-cadherin. Onco Targets Ther. 2017, 10, 1783–1792. [Google Scholar] [CrossRef]
- Ye, G.; Qin, Y.; Wang, S.; Pan, D.; Xu, S.; Wu, C.; Wang, X.; Wang, J.; Ye, H.; Shen, H. Lamc1 promotes the Warburg effect in hepatocellular carcinoma cells by regulating PKM2 expression through AKT pathway. Cancer Biol. Ther. 2019, 20, 711–719. [Google Scholar] [CrossRef] [Green Version]
- Patarroyo, M.; Tryggvason, K.; Virtanen, I. Laminin isoforms in tumor invasion, angiogenesis and metastasis. Semin. Cancer Biol. 2002, 12, 197–207. [Google Scholar] [CrossRef]
- Gilkes, D.M.; Bajpai, S.; Chaturvedi, P.; Wirtz, D.; Semenza, G.L. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J. Biol. Chem. 2013, 288, 10819–10829. [Google Scholar] [CrossRef] [PubMed]
- Victor, B.C.; Anbalagan, A.; Mohamed, M.M.; Sloane, B.F.; Cavallo-Medved, D. Inhibition of cathepsin B activity attenuates extracellular matrix degradation and inflammatory breast cancer invasion. Breast Cancer Res. 2011, 13, R115. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, N.S.; Nagaraj, N.S.; Vigneswaran, N.; Zacharias, W. Cathepsin B promotes both motility and invasiveness of oral carcinoma cells. Arch. Biochem. Biophys. 2005, 436, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Givant-Horwitz, V.; Davidson, B.; Reich, R. Laminin-induced signaling in tumor cells. Cancer Lett. 2005, 223, 1–10. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Xu, Q.; Niu, X.; Wang, W.; Yang, W.; Du, Y.; Gu, J.; Song, L. Specific N-glycan alterations are coupled in EMT induced by different density cultivation of MCF 10A epithelial cells. Glycoconj. J. 2017, 34, 219–227. [Google Scholar] [CrossRef]
- Tzanakakis, G.; Kavasi, R.M.; Voudouri, K.; Berdiaki, A.; Spyridaki, I.; Tsatsakis, A.; Nikitovic, D. Role of the extracellular matrix in cancer-associated epithelial to mesenchymal transition phenomenon. Dev. Dyn. 2018, 247, 368–381. [Google Scholar] [CrossRef]
- Jolly, M.K.; Somarelli, J.A.; Sheth, M.; Biddle, A.; Tripathi, S.C.; Armstrong, A.J.; Hanash, S.M.; Bapat, S.A.; Rangarajan, A.; Levine, H. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol. Ther. 2019, 194, 161–184. [Google Scholar] [CrossRef]




| Gene Symbol | Glycosylation Site | Glycosylation Fold Change | Total Protein Fold Change * |
|---|---|---|---|
| LAMC1 | 3915 | 2.0 | 1.1 |
| SERPINH1 | 120 | 2.8 | 1.5 |
| PLOD2 | 5352 | 3.0 | 1.8 |
| TIMP1 | 7076 | 1.9 | NI |
| PLOD1 | 5351 | 1.8 | NI |
| LAMA3 | 3909 | 0.7 | NI |
| P4HA1 | 5033 | 2.7 | 1.3 |
| LAMA4 | 3910 | 2.0 | NI |
| CTSB | 1508 | 2.1 | 1.3 |
| ITGA1 | 217 | 1.6 | NI |
| LAMA5 | 3911 | 0.7 | NI |
| CTSD | 263 | 0.5 | 0.7 |
| ITGB6 | 97 | 0.3 | NI |
| ITGA5 | 3678 | 3.2 | 1.8 |
| FN1 | 2335 | 2.0 | 2.2 |
| NCAM1 | 485 | 2.3 | NI |
| ICAM1 | 260 | 0.4 | 0.7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deb, B.; Patel, K.; Sathe, G.; Kumar, P. N-Glycoproteomic Profiling Reveals Alteration In Extracellular Matrix Organization In Non-Type Bladder Carcinoma. J. Clin. Med. 2019, 8, 1303. https://doi.org/10.3390/jcm8091303
Deb B, Patel K, Sathe G, Kumar P. N-Glycoproteomic Profiling Reveals Alteration In Extracellular Matrix Organization In Non-Type Bladder Carcinoma. Journal of Clinical Medicine. 2019; 8(9):1303. https://doi.org/10.3390/jcm8091303
Chicago/Turabian StyleDeb, Barnali, Krishna Patel, Gajanan Sathe, and Prashant Kumar. 2019. "N-Glycoproteomic Profiling Reveals Alteration In Extracellular Matrix Organization In Non-Type Bladder Carcinoma" Journal of Clinical Medicine 8, no. 9: 1303. https://doi.org/10.3390/jcm8091303
APA StyleDeb, B., Patel, K., Sathe, G., & Kumar, P. (2019). N-Glycoproteomic Profiling Reveals Alteration In Extracellular Matrix Organization In Non-Type Bladder Carcinoma. Journal of Clinical Medicine, 8(9), 1303. https://doi.org/10.3390/jcm8091303