Genome-Wide Copy Number Variation Association Study of Atrial Fibrillation Related Thromboembolic Stroke
Abstract
:1. Introduction
2. Methods
2.1. Study Population and Outcome Assessments
2.2. DNA Extraction and Genome-Wide Detection of CNV
2.3. Pathway Analysis
2.4. Statistical Analysis
3. Results
3.1. General CNV Pattern and Association with the Risk of Thromboembolic Stroke Induced by AF
3.2. Functional Integration of Identified CNVs and Genes by Pathway Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kakar, P.; Boos, C.J.; Lip, G.Y. Management of atrial fibrillation. Vasc. Health Risk Manag. 2007, 3, 109–116. [Google Scholar] [PubMed]
- Crandall, M.A.; Horne, B.D.; Day, J.D.; Anderson, J.L.; Muhlestein, J.B.; Crandall, B.G.; Weiss, J.P.; Lappe, D.L.; Bunch, T.J. Atrial fibrillation and CHADS2 risk factors are associated with highly sensitive C-reactive protein incrementally and independently. Pacing Clin. Electrophysiol. 2009, 32, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Abraham, J.M.; Larson, J.; Chung, M.K.; Curtis, A.B.; Lakshminarayan, K.; Newman, J.D.; Perez, M.; Rexrode, K.; Shara, N.M.; Solomon, A.J.; et al. Does CHA2DS2-VASc improve stroke risk stratification in postmenopausal women with atrial fibrillation? Am. J. Med. 2013, 126, 1143–1148. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.N.; Lai, L.P.; Chiang, F.T.; Lin, J.L.; Hwang, J.J.; Tsai, C.T. C-reactive protein gene polymorphism predicts the risk of thromboembolic stroke in patients with atrial fibrillation: A more than 10-year prospective follow-up study. J. Thromb. Haemost. 2017, 15, 1541–1546. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.T.; Chang, S.N.; Chang, S.H.; Lee, J.K.; Lin, L.Y.; Wu, C.K.; Yu, C.C.; Wang, Y.C.; Tseng, C.D.; Lai, L.P.; et al. Renin-angiotensin system gene polymorphisms predict the risk of stroke in patients with atrial fibrillation: A 10-year prospective follow-up study. Heart Rhythm Off. J. Heart Rhythm Soc. 2014, 11, 1384–1390. [Google Scholar] [CrossRef] [PubMed]
- Stranger, B.E.; Forrest, M.S.; Dunning, M.; Ingle, C.E.; Beazley, C.; Thorne, N.; Redon, R.; Bird, C.P.; de Grassi, A.; Lee, C.; et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Sciences 2007, 315, 848–853. [Google Scholar] [CrossRef]
- Conrad, D.F.; Pinto, D.; Redon, R.; Feuk, L.; Gokcumen, O.; Zhang, Y.; Aerts, J.; Andrews, T.D.; Barnes, C.; Campbell, P.; et al. Origins and functional impact of copy number variation in the human genome. Nature 2010, 464, 704–712. [Google Scholar] [CrossRef]
- Lanktree, M.; Hegele, R.A. Copy number variation in metabolic phenotypes. Cytogenet. Genome Res. 2008, 123, 169–175. [Google Scholar] [CrossRef]
- Chen, C.H.; Yang, J.H.; Chiang, C.W.K.; Hsiung, C.N.; Wu, P.E.; Chang, L.C.; Chu, H.W.; Chang, J.; Song, I.W.; Yang, S.L.; et al. Population structure of Han Chinese in the modern Taiwanese population based on 10,000 participants in the Taiwan Biobank project. Hum. Mol. Genet. 2016, 25, 5321–5331. [Google Scholar] [CrossRef]
- Wu, C.K.; Juang, J.J.; Chiang, J.Y.; Li, Y.H.; Tsai, C.T.; Chiang, F.T. The Taiwan Heart Registries: Its Influence on Cardiovascular Patient Care. J. Am. Coll. Cardiol. 2018, 71, 1273–1283. [Google Scholar] [CrossRef]
- Lahiri, D.K.; Nurnberger, J.I., Jr. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991, 19, 5444. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.P.; Lai, L.C.; Tsai, M.H.; Chen, P.C.; Hsu, C.P.; Lee, J.M.; Hsiao, C.K.; Chuang, E.Y. Integrated analyses of copy number variations and gene expression in lung adenocarcinoma. PLoS ONE 2011, 6, e24829. [Google Scholar] [CrossRef] [PubMed]
- Shia, W.C.; Ku, T.H.; Tsao, Y.M.; Hsia, C.H.; Chang, Y.M.; Huang, C.H.; Chung, Y.C.; Hsu, S.L.; Liang, K.W.; Hsu, F.R. Genetic copy number variants in myocardial infarction patients with hyperlipidemia. BMC Genom. 2011, 12 (Suppl. 3), S23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Tsvetanova, N.G.; Irannejad, R.; von Zastrow, M. G protein-coupled receptor (GPCR) signaling via heterotrimeric G proteins from endosomes. J. Biol. Chem. 2015, 290, 6689–6696. [Google Scholar] [CrossRef] [PubMed]
- Nattel, S.; Burstein, B.; Dobrev, D. Atrial remodeling and atrial fibrillation: Mechanisms and implications. Circ. Arrhythmia Electrophysiol. 2008, 1, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Tsao, H.M.; Hu, W.C.; Tsai, P.H.; Lee, C.L.; Wang, H.H.; Chang, S.L.; Chao, T.F.; Chen, S.A. Functional Remodeling of Both Atria is Associated with Occurrence of Stroke in Patients with Paroxysmal and Persistent Atrial Fibrillation. Acta Cardiol. Sin. 2017, 33, 50–57. [Google Scholar]
- Vatan, M.B.; Yilmaz, S.; Agac, M.T.; Cakar, M.A.; Erkan, H.; Aksoy, M.; Demirtas, S.; Varim, C.; Akdemir, R.; Gunduz, H. Relationship between CHA2DS2-VASc score and atrial electromechanical function in patients with paroxysmal atrial fibrillation: A pilot study. J. Cardiol. 2015, 66, 382–387. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Gaspo, R.; Leblanc, N.; Nattel, S. Cellular mechanisms of atrial contractile dysfunction caused by sustained atrial tachycardia. Circulation 1998, 98, 719–727. [Google Scholar] [CrossRef]
- Yue, L.; Feng, J.; Gaspo, R.; Li, G.R.; Wang, Z.; Nattel, S. Ionic remodeling underlying action potential changes in a canine model of atrial fibrillation. Circ. Res. 1997, 81, 512–525. [Google Scholar] [CrossRef]
- Christ, T.; Boknik, P.; Wohrl, S.; Wettwer, E.; Graf, E.M.; Bosch, R.F.; Knaut, M.; Schmitz, W.; Ravens, U.; Dobrev, D. L-type Ca2+ current downregulation in chronic human atrial fibrillation is associated with increased activity of protein phosphatases. Circulation 2004, 110, 2651–2657. [Google Scholar] [CrossRef] [PubMed]
- Greiser, M.; Halaszovich, C.R.; Frechen, D.; Boknik, P.; Ravens, U.; Dobrev, D.; Luckhoff, A.; Schotten, U. Pharmacological evidence for altered src kinase regulation of I (Ca,L) in patients with chronic atrial fibrillation. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2007, 375, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Christ, T.; Rozmaritsa, N.; Engel, A.; Berk, E.; Knaut, M.; Metzner, K.; Canteras, M.; Ravens, U.; Kaumann, A. Arrhythmias, elicited by catecholamines and serotonin, vanish in human chronic atrial fibrillation. Proc. Natl. Acad. Sci. USA 2014, 111, 11193–11198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuldeep, C.M.; Khare, A.K.; Garg, A.; Mittal, A.; Gupta, L. Terminal 4q deletion syndrome. Indian J. Dermatol. 2012, 57, 222–224. [Google Scholar] [CrossRef] [PubMed]
- Strehle, E.M.; Yu, L.; Rosenfeld, J.A.; Donkervoort, S.; Zhou, Y.; Chen, T.J.; Martinez, J.E.; Fan, Y.S.; Barbouth, D.; Zhu, H.; et al. Genotype-phenotype analysis of 4q deletion syndrome: Proposal of a critical region. Am. J. Med. Genet. Part A 2012, 158, 2139–2151. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Ahmad, A.; Dagenais, S.; Iyer, R.K.; Innis, J.W. Chromosome 4q deletion syndrome: Narrowing the cardiovascular critical region to 4q32.2-q34.3. Am. J. Med. Genet. Part A 2012, 158, 635–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peddibhotla, S.; Nagamani, S.C.; Erez, A.; Hunter, J.V.; Holder, J.L., Jr.; Carlin, M.E.; Bader, P.I.; Perras, H.M.; Allanson, J.E.; Newman, L.; et al. Delineation of candidate genes responsible for structural brain abnormalities in patients with terminal deletions of chromosome 6q27. Eur. J. Hum. Genet. 2015, 23, 54–60. [Google Scholar] [CrossRef]
- Su, P.H.; Chen, J.Y.; Chen, S.J.; Yang, K.C. Terminal deletion of chromosome 6q. Pediatr. Neonatol. 2008, 49, 88–93. [Google Scholar] [CrossRef]
- Tsai, C.T.; Hsieh, C.S.; Chang, S.N.; Chuang, E.Y.; Ueng, K.C.; Tsai, C.F.; Lin, T.H.; Wu, C.K.; Lee, J.K.; Lin, L.Y.; et al. Genome-wide screening identifies a KCNIP1 copy number variant as a genetic predictor for atrial fibrillation. Nat. Commun. 2016, 7, 10190. [Google Scholar] [CrossRef] [Green Version]

| Variables | AF with Stroke (n = 109) | Controls (n = 14,666) |
|---|---|---|
| Age (yr) * | 71 ± 12 | 48 ± 11 |
| Male (%) | 53 (48.6) | 7246 (49.4) |
| DM (%) | 9 (8.3) | 656 (4.4) |
| HTN (%) | 18 (16.5) | 1589 (10.8) |
| HPL (%) | 11 (10.1) | 848 (5.7) |
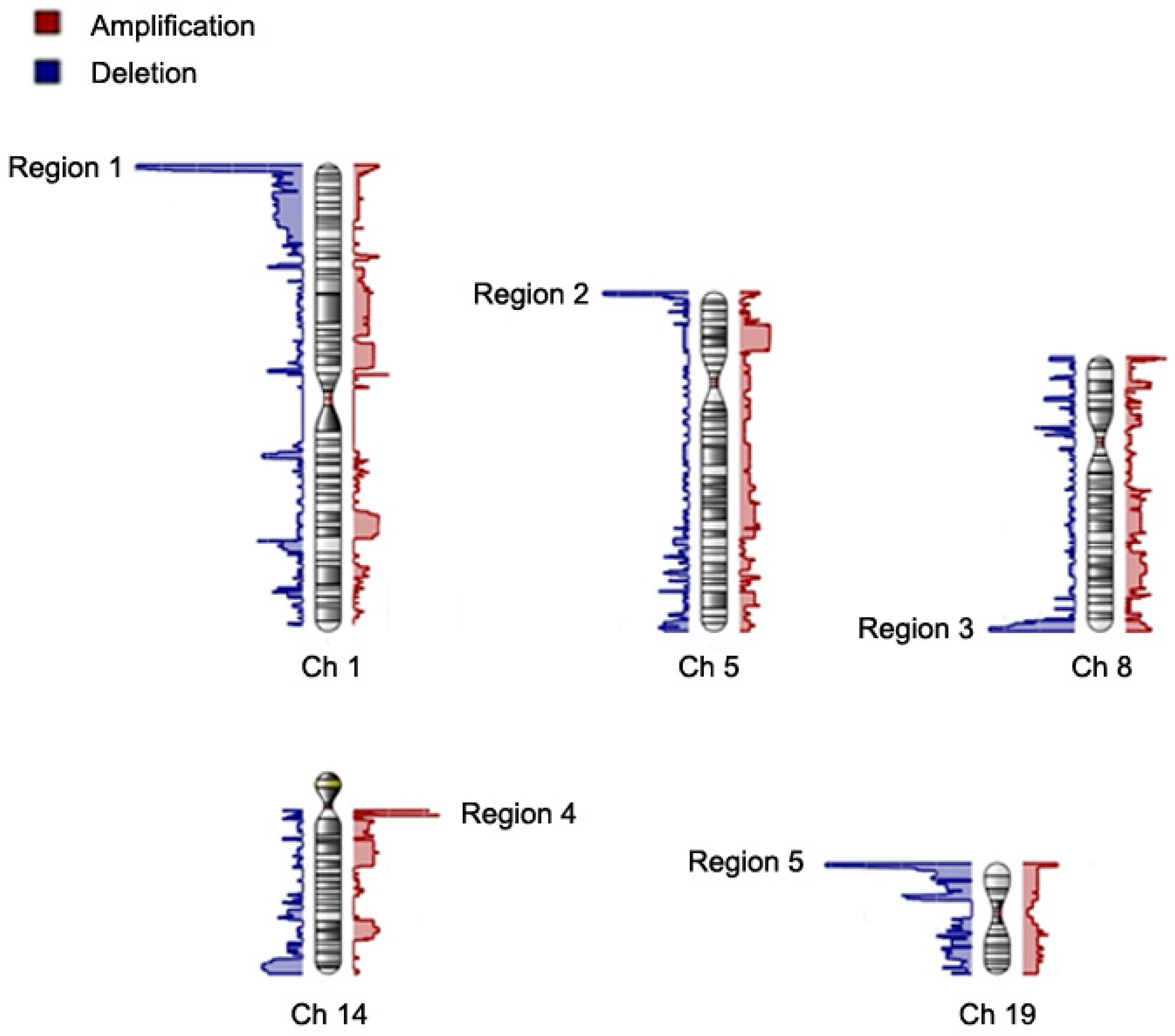
| CNV Region | Cytoband | Start Position | Type | Allele Frequency | Avg CN | Length (bps) | Nearest Feature | p | ln(OR) (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| Region 1 | 1p36.33 | 565433 | Del | 0.30 | 1.45 | 1,614,263 | GNB1, PRKCZ | 1.69 × 10−10 | 15.3 (12.9–17.7) |
| 1p36.33-1p36.32 | 2179695 | Del | 0.30 | 1.45 | 298,359 | RP3-395M20.12 | 2.14 × 10−11 | 17.5 (14.9–20.1) | |
| 1p36.32 | 2478053 | Del | 0.28 | 1.44 | 391,244 | RP11-740P5.3 | 2.95 × 10−9 | 15.8 (13.1–18.4) | |
| 1p36.32 | 2869296 | Del | 0.27 | 1.43 | 293,652 | PRDM16 | 6.24 × 10−9- | 13.8 (11.5–16.2) | |
| 1p36.32 | 3162947 | Del | 0.26 | 1.42 | 10,249 | PRDM16 | 8.58 × 10−9 | 13.8 (11.4–16.2) | |
| 1p36.32 | 3173195 | Del | 0.24 | 1.41 | 184,908 | PRDM16 | 1.44 × 10−8 | 13.8 (11.3–16.2) | |
| 1p36.32 | 3358102 | Del | 0.24 | 1.41 | 297,333 | TP73-AS1 | 5.09 × 10−9 | 15.8 (13.1–18.5) | |
| 1p36.32 | 3655434 | Del | 0.23 | 1.41 | 7371 | TP73-AS1 | 5.53 × 10−9 | 15.8 (13.1–18.5) | |
| 1p36.32 | 3662804 | Del | 0.22 | 1.41 | 18,596 | CCDC27 | 3.44 × 10−7 | 14.4 (11.6–17.2) | |
| 1p36.32 | 3681399 | Del | 0.21 | 1.40 | 11,306 | SMIM1 | 3.86 × 10−7 | 14.4 (11.6–17.3) | |
| 1p36.32 | 3692704 | Del | 0.20 | 1.39 | 3565 | SMIM1 | 5.03 × 10−7 | 14.4 (11.5–17.2) | |
| 1p36.32 | 3696268 | Del | 0.19 | 1.38 | 45,605 | CEP104 | 5.74 × 10−7 | 14.4 (11.5–17.3) | |
| 1p36.32 | 3741872 | Del | 0.17 | 1.36 | 58,040 | DFFB | 1.2 × 10−5 | 11.9 (9.2–14.7) | |
| 1p36.32 | 3799911 | Del | 0.17 | 1.36 | 205,245 | RP13-614K11.1 | 1.53 × 10−5 | 11.9 (9.2–14.7) | |
| Region 2 | 5p15.33 | 1313855 | Del | 0.16 | 1.39 | 158,260 | LPCAT1 | 1.9 × 10−5 | 10.0 (7.7–12.4) |
| 5p15.33 | 1472114 | Del | 0.16 | 1.39 | 193,377 | RP11-43F13.1 | 5.24 × 10−5 | 13.3 (10.0–16.6) | |
| Region 3 | 8q24.3 | 144283121 | Del | 0.16 | 1.43 | 357,744 | GSDMD | 5.56 × 10−6 | 8.5 (6.6–10.4) |
| Region 4 | 14q11.2 | 22754700 | Amp | 0.15 | 2.45 | 1721 | TRAV38-2DV8 | 8.23 × 10−7 | 12.6 (10.0–15.1) |
| 14q11.2 | 22756420 | Amp | 0.16 | 2.46 | 178,718 | TRDC | 1.79 × 10−9 | 14.6 (12.1–17.0) | |
| 14q11.2 | 22935137 | Amp | 0.15 | 2.46 | 7248 | TRDV3 | 1.68 × 10−8 | 14.2 (11.7–16.7) | |
| Region 5 | 19p13.3 | 2089647 | Del | 0.20 | 1.30 | 38,105 | AP3D1 | 1.64 × 10−6 | 6.9 (5.5–8.3) |
| 19p13.3 | 2127751 | Del | 0.20 | 1.34 | 23,119 | AP3D1 | 6.25 × 10−7 | 8.2 (6.5–9.8) | |
| 19p13.3 | 2150869 | Del | 0.19 | 1.35 | 19,047 | DOT1L | 3.97 × 10−8 | 11 (9.0–13.0) | |
| 19p13.3 | 2169915 | Del | 0.19 | 1.35 | 945 | DOT1L | 3.33 × 10−7 | 7.4 (6.0–8.9) | |
| 19p13.3 | 2170859 | Del | 0.19 | 1.35 | 139,329 | LINGO3 | 2.2 × 10−7 | 8.4 (6.8–10.5) | |
| 19p13.3 | 2310187 | Del | 0.19 | 1.36 | 4407 | LSM7 | 3.57 × 10−8 | 11.0 (9.0–13.0) | |
| 19p13.3 | 2314593 | Del | 0.17 | 1.35 | 86,339 | TMPRSS9 | 1.44 × 10−6 | 9.9 (7.8–11.9) | |
| 19p13.3 | 2400931 | Del | 0.17 | 1.33 | 3631 | TMPRSS9 | 2.7 × 10−6 | 9.7 (7.7–11.8) | |
| 19p13.3 | 2404561 | Del | 0.16 | 1.32 | 73,270 | GADD45B | 6.5 × 10−6 | 9.5 (7.4–11.7) | |
| 19p13.3 | 2477830 | Del | 0.15 | 1.32 | 30,337 | RNU6-993P | 8.82 × 10−6 | 9.5 (7.4–11.7) | |
| 19p13.3 | 2508166 | Del | 0.15 | 1.34 | 21,100 | GNG7 | 3.21 × 10−6 | 10 (7.8–12.1) |
| Gene Symbol | Cytoband | Entrez Gene Name | Location | Type(s) |
|---|---|---|---|---|
| GNB1 | 1p36.32 | G protein subunit beta 1 | Plasma Membrane | enzyme |
| GNG7 | 19p13.3 | G protein subunit gamma 7 | Plasma Membrane | enzyme |
| PRKCZ | 1p36.32 | protein kinase C zeta | Cytoplasm | kinase |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsieh, C.-S.; Huang, P.-S.; Chang, S.-N.; Wu, C.-K.; Hwang, J.-J.; Chuang, E.Y.; Tsai, C.-T. Genome-Wide Copy Number Variation Association Study of Atrial Fibrillation Related Thromboembolic Stroke. J. Clin. Med. 2019, 8, 332. https://doi.org/10.3390/jcm8030332
Hsieh C-S, Huang P-S, Chang S-N, Wu C-K, Hwang J-J, Chuang EY, Tsai C-T. Genome-Wide Copy Number Variation Association Study of Atrial Fibrillation Related Thromboembolic Stroke. Journal of Clinical Medicine. 2019; 8(3):332. https://doi.org/10.3390/jcm8030332
Chicago/Turabian StyleHsieh, Chia-Shan, Pang-Shuo Huang, Sheng-Nan Chang, Cho-Kai Wu, Juey-Jen Hwang, Eric Y. Chuang, and Chia-Ti Tsai. 2019. "Genome-Wide Copy Number Variation Association Study of Atrial Fibrillation Related Thromboembolic Stroke" Journal of Clinical Medicine 8, no. 3: 332. https://doi.org/10.3390/jcm8030332




