MiR-122 Targets SerpinB3 and Is Involved in Sorafenib Resistance in Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Cell Lines
2.3. Luciferase Assay
2.4. DEN-HCC Rat Model
2.5. Real-Time PCR
2.6. Western Blot
2.7. MiR-122 In Situ Hybridization (ISH) and Immunohistochemical (IHC) Analysis
2.8. Flow Cytometry
2.9. Statistical Analysis
3. Results
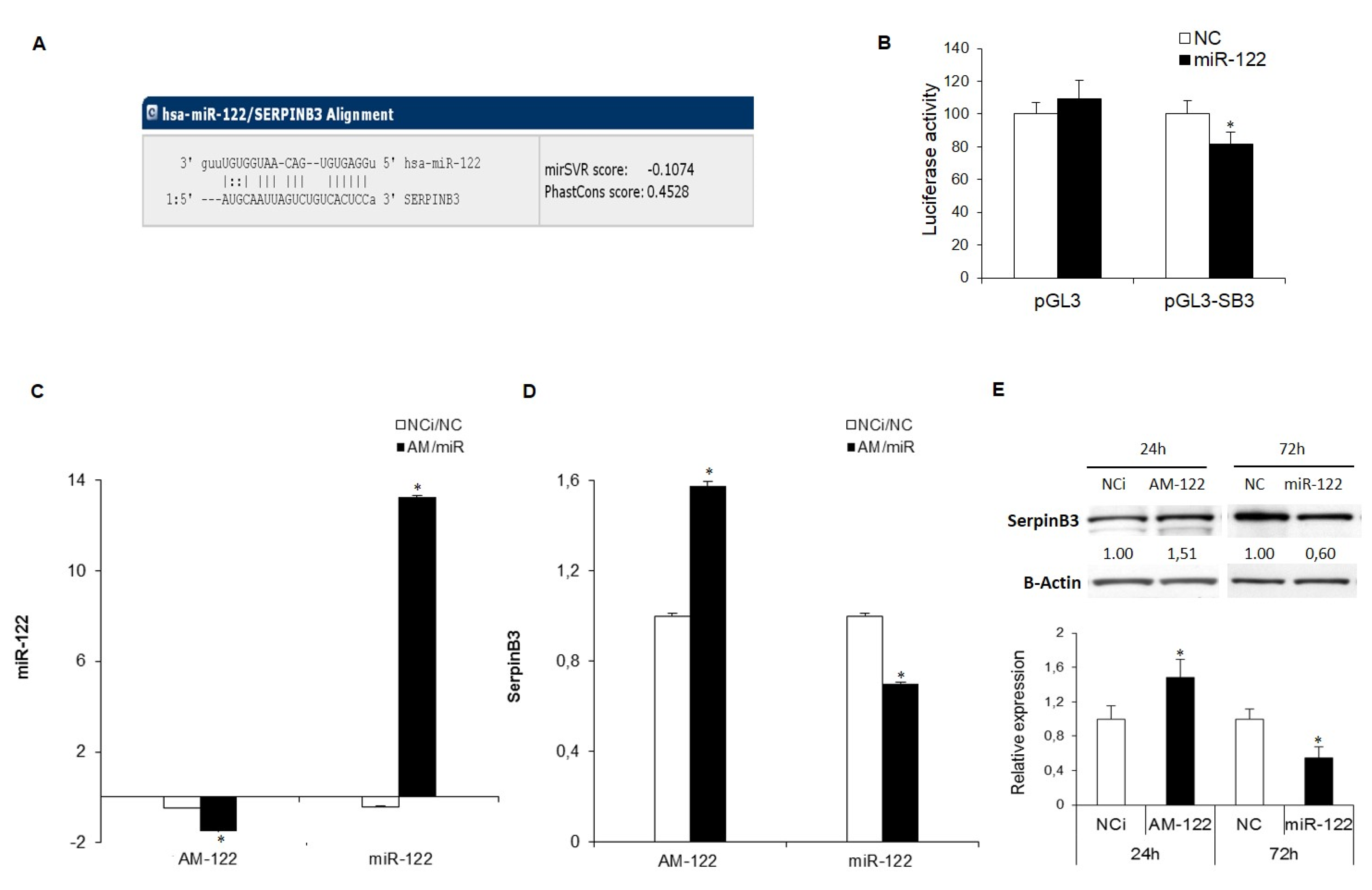
3.1. SerpinB3 Is a Target of miR-122 in HCC
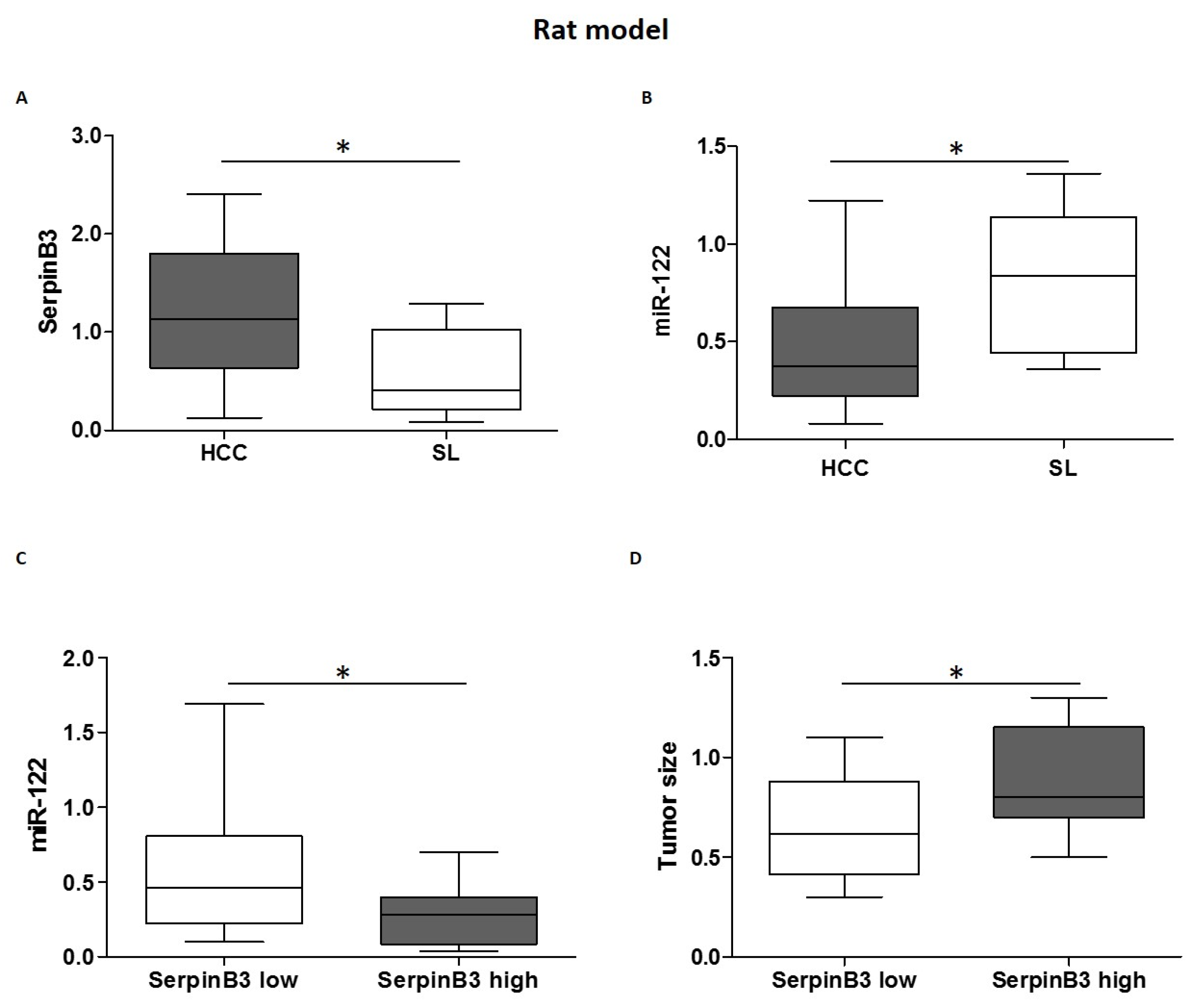
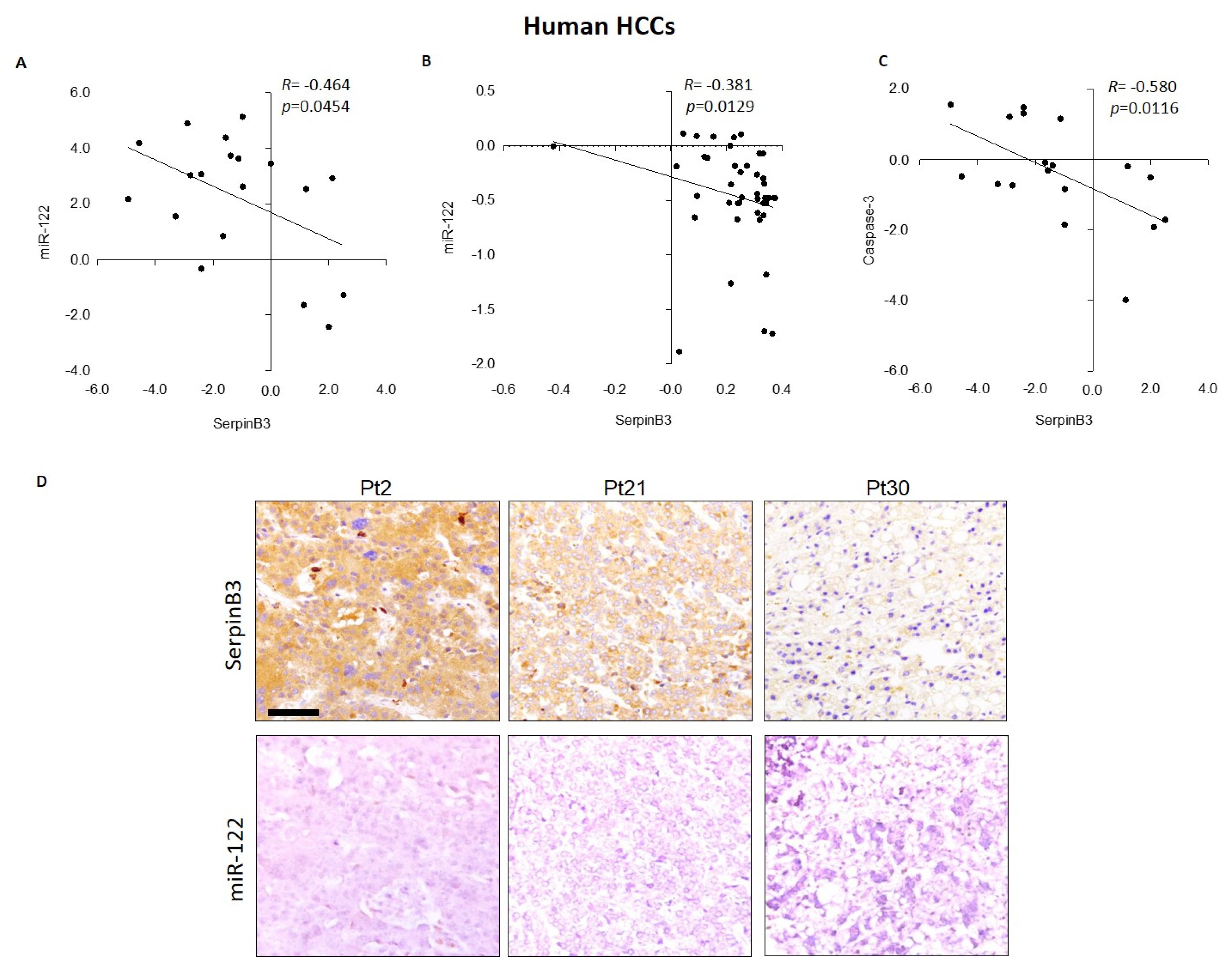
3.2. MiR-122 and SerpinB3 Inversely Correlate In Vivo
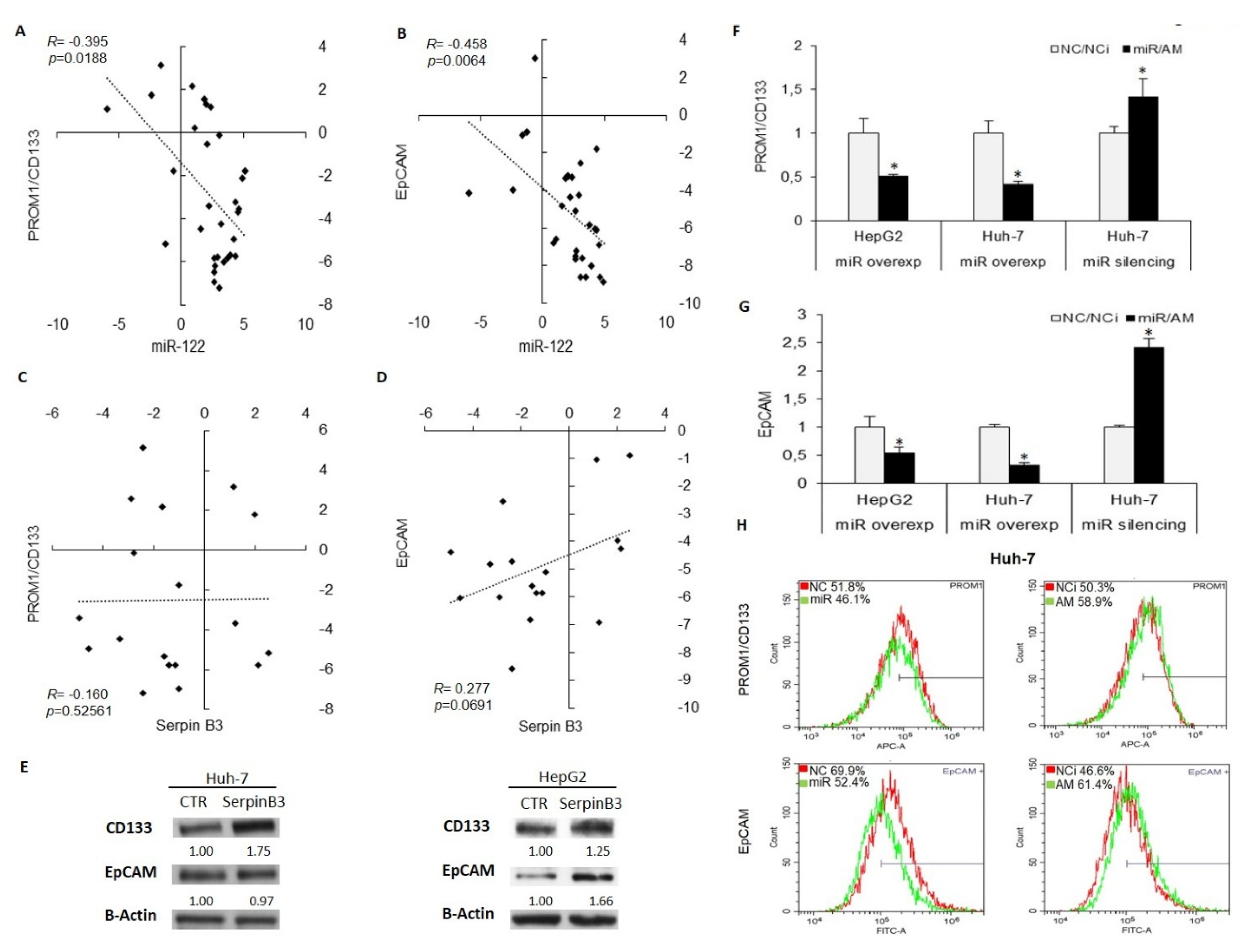
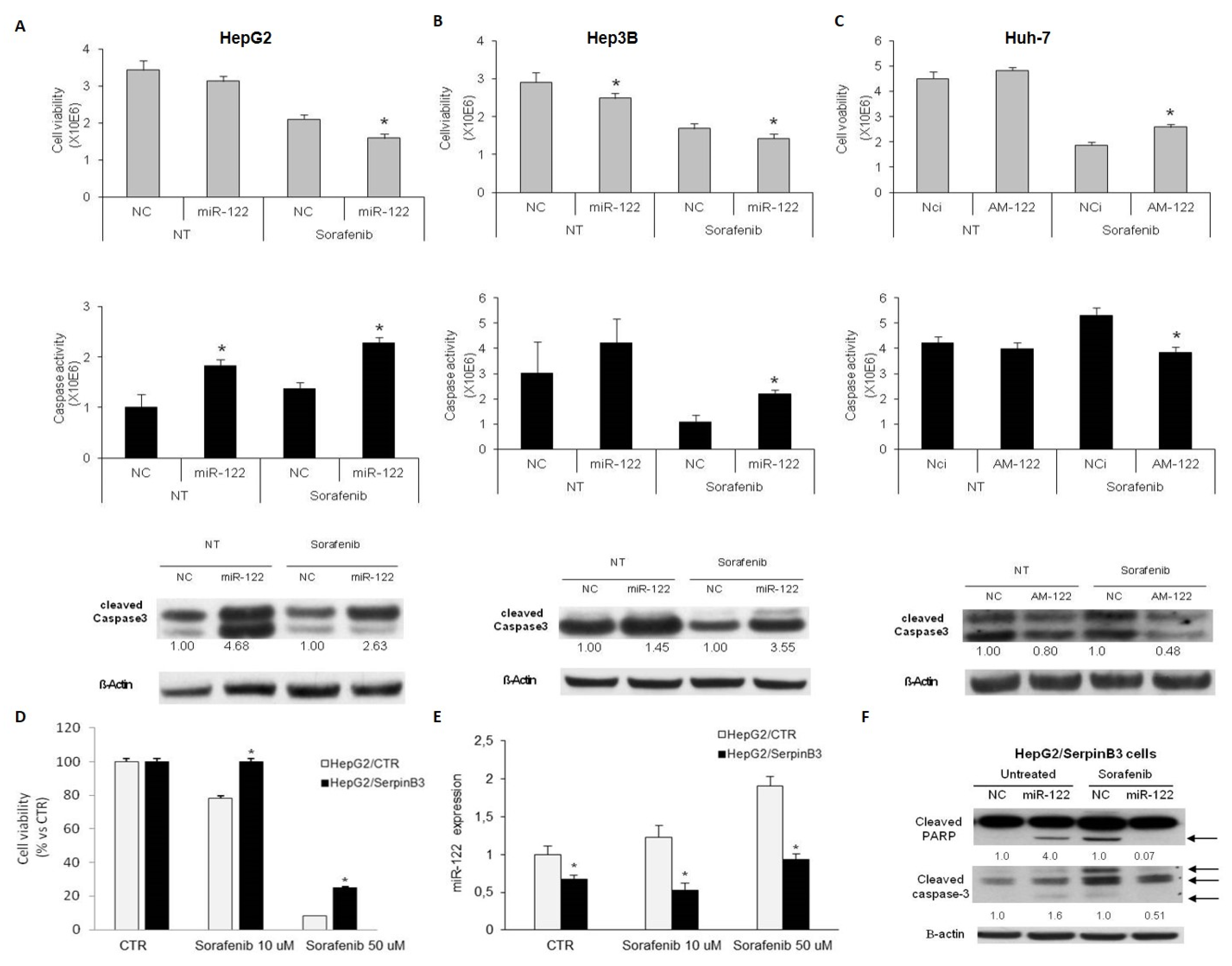
3.3. MiR-122 and SerpinB3 Modulate Sorafenib Response in HCC Cell Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2016, 2, 16018. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. SHARP Investigators Study Group sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—Micrornas with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Rao, D.S.; Baltimore, D. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012, 30, 295–312. [Google Scholar] [CrossRef]
- Szabo, G.; Bala, S. MicroRNAs in liver disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Thakral, S.; Ghoshal, K. MiR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr. Gene. Ther. 2015, 15, 142–150. [Google Scholar] [CrossRef]
- Castoldi, M.; Vujic Spasic, M.; Altamura, S.; Elmen, J.; Lindow, M.; Kiss, J.; Stolte, J.; Sparla, R.; D’Alessandro, L.A.; Klingmuller, U.; et al. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J. Clin. Investig. 2011, 121, 1386–1396. [Google Scholar] [CrossRef]
- Fan, B.; Sutandy, F.X.; Syu, G.D.; Middleton, S.; Yi, G.; Lu, K.Y.; Chen, C.S.; Kao, C.C. Heterogeneous Ribonucleoprotein K (hnRNP K) Binds miR-122, a Mature Liver-Specific MicroRNA Required for Hepatitis C Virus Replication. Mol. Cell. Proteom. 2015, 14, 2878–2886. [Google Scholar] [CrossRef]
- Coulouarn, C.; Factor, V.M.; Andersen, J.B.; Durkin, M.E.; Thorgeirsson, S.S. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 2009, 28, 3526–3536. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.C.; Hsu, P.W.; Lai, T.C.; Chau, G.Y.; Lin, C.W.; Chen, C.M.; Lin, C.D.; Liao, Y.L.; Wang, J.L.; Chau, Y.P.; et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology 2009, 49, 1571–1582. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.H.; Wang, B.; Kota, J.; Yu, J.; Costinean, S.; Kutay, H.; Yu, L.; Bai, S.; La Perle, K.; Chivukula, R.R.; et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Investig. 2012, 122, 2871–2883. [Google Scholar] [CrossRef] [PubMed]
- Gramantieri, L.; Ferracin, M.; Fornari, F.; Veronese, A.; Sabbioni, S.; Liu, C.G.; Calin, G.A.; Giovannini, C.; Ferrazzi, E.; Grazi, G.L.; et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007, 67, 6092–6099. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; He, J.H.; Xiao, Z.D.; Zhang, Q.Q.; Chen, Y.Q.; Zhou, H.; Qu, L.H. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology 2010, 52, 1431–1442. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xia, F.; Ma, L.; Shan, J.; Shen, J.; Yang, Z.; Liu, J.; Cui, Y.; Bian, X.; Bie, P.; et al. MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. 2011, 310, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Turato, C.; Simonato, D.; Quarta, S.; Gatta, A.; Pontisso, P. MicroRNAs and SerpinB3 in hepatocellular carcinoma. Life Sci. 2014, 100, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Guido, M.; Roskams, T.; Pontisso, P.; Fassan, M.; Thung, S.N.; Giacomelli, L.; Sergio, A.; Farinati, F.; Cillo, U.; Rugge, M. Squamous cell carcinoma antigen in human liver carcinogenesis. J. Clin. Pathol. 2008, 61, 445–447. [Google Scholar] [CrossRef] [PubMed]
- Turato, C.; Vitale, A.; Fasolato, S.; Ruvoletto, M.; Terrin, L.; Quarta, S.; Ramirez Morales, R.; Biasiolo, A.; Zanus, G.; Zali, N.; et al. SERPINB3 is associated with TGF-beta1 and cytoplasmic beta-catenin expression in hepatocellular carcinomas with poor prognosis. Br. J. Cancer 2014, 110, 2708–2715. [Google Scholar] [CrossRef] [PubMed]
- Quarta, S.; Vidalino, L.; Turato, C.; Ruvoletto, M.; Calabrese, F.; Valente, M.; Cannito, S.; Fassina, G.; Parola, M.; Gatta, A.; et al. SERPINB3 induces epithelial-mesenchymal transition. J. Pathol. 2010, 221, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Pollutri, D.; Patrizi, C.; La Bella, T.; Marinelli, S.; Casadei Gardini, A.; Marisi, G.; Baron Toaldo, M.; Baglioni, M.; Salvatore, V.; et al. In Hepatocellular Carcinoma miR-221 Modulates sorafenib Resistance through Inhibition of Caspase-3-Mediated Apoptosis. Clin. Cancer Res. 2017, 23, 3953–3965. [Google Scholar] [CrossRef] [PubMed]
- Cannito, S.; Turato, C.; Paternostro, C.; Biasiolo, A.; Colombatto, S.; Cambieri, I.; Quarta, S.; Novo, E.; Morello, E.; Villano, G.; et al. Hypoxia up-regulates SERPINB3 through HIF-2alpha in human liver cancer cells. Oncotarget 2015, 6, 2206–2221. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Gramantieri, L.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Giovannini, C.; Croce, C.M.; Bolondi, L.; et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 2008, 27, 5651–5661. [Google Scholar] [CrossRef] [PubMed]
- Turato, C.; Cannito, S.; Simonato, D.; Villano, G.; Morello, E.; Terrin, L.; Quarta, S.; Biasiolo, A.; Ruvoletto, M.; Martini, A.; et al. SerpinB3 and Yap Interplay Increases Myc Oncogenic Activity. Sci. Rep. 2015, 5, 17701. [Google Scholar] [CrossRef] [PubMed]
- Saraggi, D.; Galuppini, F.; Fanelli, G.N.; Remo, A.; Urso, E.D.L.; Bao, R.Q.; Bacchin, D.; Guzzardo, V.; Luchini, C.; Braconi, C.; et al. MiR-21 up-regulation in ampullary adenocarcinoma and its pre-invasive lesions. Pathol. Res. Pract. 2018, 214, 835–839. [Google Scholar] [CrossRef] [PubMed]
- Cui, R.; Kim, T.; Fassan, M.; Meng, W.; Sun, H.L.; Jeon, Y.J.; Vicentini, C.; Tili, E.; Peng, Y.; Scarpa, A.; et al. MicroRNA-224 is implicated in lung cancer pathogenesis through targeting caspase-3 and caspase-7. Oncotarget 2015, 6, 21802–21815. [Google Scholar] [CrossRef] [PubMed]
- Frau, M.; Simile, M.M.; Tomasi, M.L.; Demartis, M.I.; Daino, L.; Seddaiu, M.A.; Brozzetti, S.; Feo, C.F.; Massarelli, G.; Solinas, G.; et al. An expression signature of phenotypic resistance to hepatocellular carcinoma identified by cross-species gene expression analysis. Cell. Oncol. 2012, 35, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lingala, S.; Khoobyari, S.; Nolta, J.; Zern, M.A.; Wu, J. Epithelial mesenchymal transition and hedgehog signaling activation are associated with chemoresistance and invasion of hepatoma subpopulations. J. Hepatol. 2011, 55, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Gramantieri, L.; Giovannini, C.; Veronese, A.; Ferracin, M.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Croce, C.M.; Tavolari, S.; et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009, 69, 5761–5767. [Google Scholar] [CrossRef]
- Bai, S.; Nasser, M.W.; Wang, B.; Hsu, S.H.; Datta, J.; Kutay, H.; Yadav, A.; Nuovo, G.; Kumar, P.; Ghoshal, K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J. Biol. Chem. 2009, 284, 32015–32027. [Google Scholar] [CrossRef]
- Jopling, C. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012, 9, 137–142. [Google Scholar] [CrossRef]
- Pontisso, P. Role of SERPINB3 in hepatocellular carcinoma. Ann. Hepatol. 2014, 13, 722–727. [Google Scholar] [PubMed]
- Cervello, M.; Bachvarov, D.; Lampiasi, N.; Cusimano, A.; Azzolina, A.; McCubrey, J.A.; Montalto, G. Molecular mechanisms of sorafenib action in liver cancer cells. Cell Cycle 2012, 11, 2843–2855. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, L.; Wang, F.; Wang, Y.; Huo, X.S.; Yin, Y.X.; Wang, Y.Q.; Zhang, L.; Sun, S.H. Modulation of the unfolded protein response is the core of microRNA-122-involved sensitivity to chemotherapy in hepatocellular carcinoma. Neoplasia 2011, 13, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Bandiera, S.; Pfeffer, S.; Baumert, T.F.; Zeisel, M.B. miR-122—A key factor and therapeutic target in liver disease. J. Hepatol. 2015, 62, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Villano, G.; Turato, C.; Quarta, S.; Ruvoletto, M.; Ciscato, F.; Terrin, L.; Semeraro, R.; Paternostro, C.; Parola, M.; Alvaro, D.; et al. Hepatic progenitor cells express SerpinB3. BMC Cell Biol. 2014, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Kwon, H.; Han, C.; Zhang, J.; Dash, S.; Lim, K.; Wu, T. Active glycolytic metabolism in CD133(+) hepatocellular cancer stem cells: Regulation by MIR-122. Oncotarget 2015, 6, 40822–40835. [Google Scholar] [CrossRef] [PubMed]
- Bimonte, S.; Leongito, M.; Barbieri, A.; Del Vecchio, V.; Falco, M.; Giudice, A.; Palaia, R.; Albino, V.; Di Giacomo, R.; Petrillo, A.; et al. The Therapeutic Targets of miRNA in Hepatic Cancer Stem Cells. Stem. Cells Int. 2016, 2016, 1065230. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.K.; Cheung, V.C.; Ng, I.O. Liver tumor-initiating cells as a therapeutic target for hepatocellular carcinoma. Cancer Lett. 2013, 338, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huang, J.; Ma, L.; Shan, J.; Shen, J.; Yang, Z.; Liu, L.; Luo, Y.; Yao, C.; Qian, C. MicroRNA-122 confers sorafenib resistance to hepatocellular carcinoma cells by targeting IGF-1R to regulate RAS/RAF/ERK signaling pathways. Cancer Lett. 2016, 371, 171–181. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turato, C.; Fornari, F.; Pollutri, D.; Fassan, M.; Quarta, S.; Villano, G.; Ruvoletto, M.; Bolondi, L.; Gramantieri, L.; Pontisso, P. MiR-122 Targets SerpinB3 and Is Involved in Sorafenib Resistance in Hepatocellular Carcinoma. J. Clin. Med. 2019, 8, 171. https://doi.org/10.3390/jcm8020171
Turato C, Fornari F, Pollutri D, Fassan M, Quarta S, Villano G, Ruvoletto M, Bolondi L, Gramantieri L, Pontisso P. MiR-122 Targets SerpinB3 and Is Involved in Sorafenib Resistance in Hepatocellular Carcinoma. Journal of Clinical Medicine. 2019; 8(2):171. https://doi.org/10.3390/jcm8020171
Chicago/Turabian StyleTurato, Cristian, Francesca Fornari, Daniela Pollutri, Matteo Fassan, Santina Quarta, Gianmarco Villano, Mariagrazia Ruvoletto, Luigi Bolondi, Laura Gramantieri, and Patrizia Pontisso. 2019. "MiR-122 Targets SerpinB3 and Is Involved in Sorafenib Resistance in Hepatocellular Carcinoma" Journal of Clinical Medicine 8, no. 2: 171. https://doi.org/10.3390/jcm8020171






