Quantification of HPV16 E6/E7 mRNA Spliced Isoforms Viral Load as a Novel Diagnostic Tool for Improving Cervical Cancer Screening
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Patients
2.3. Uterine Cervical Samples Analysis and Criteria of Selection
2.4. HR-HPV DNA Genotyping
2.5. HR-HPV E6/E7 mRNA Detection
2.6. Nucleic Acid Extractions
2.7. HPV16 DNA VL Quantification Assay
2.8. HPV16 E6/E7 mRNA VLs Quantification Assay
2.9. Statistical Analysis
3. Results
3.1. Validation of HPV16 DNA and E6/E7 mRNA VLs Quantification Assays
3.2. Quantification of HPV16 DNA and E6/E7 mRNA VLs in UCS
3.3. HPV16 E6/E7 mRNA VLs Are Increased in High Grade Cervical Lesions
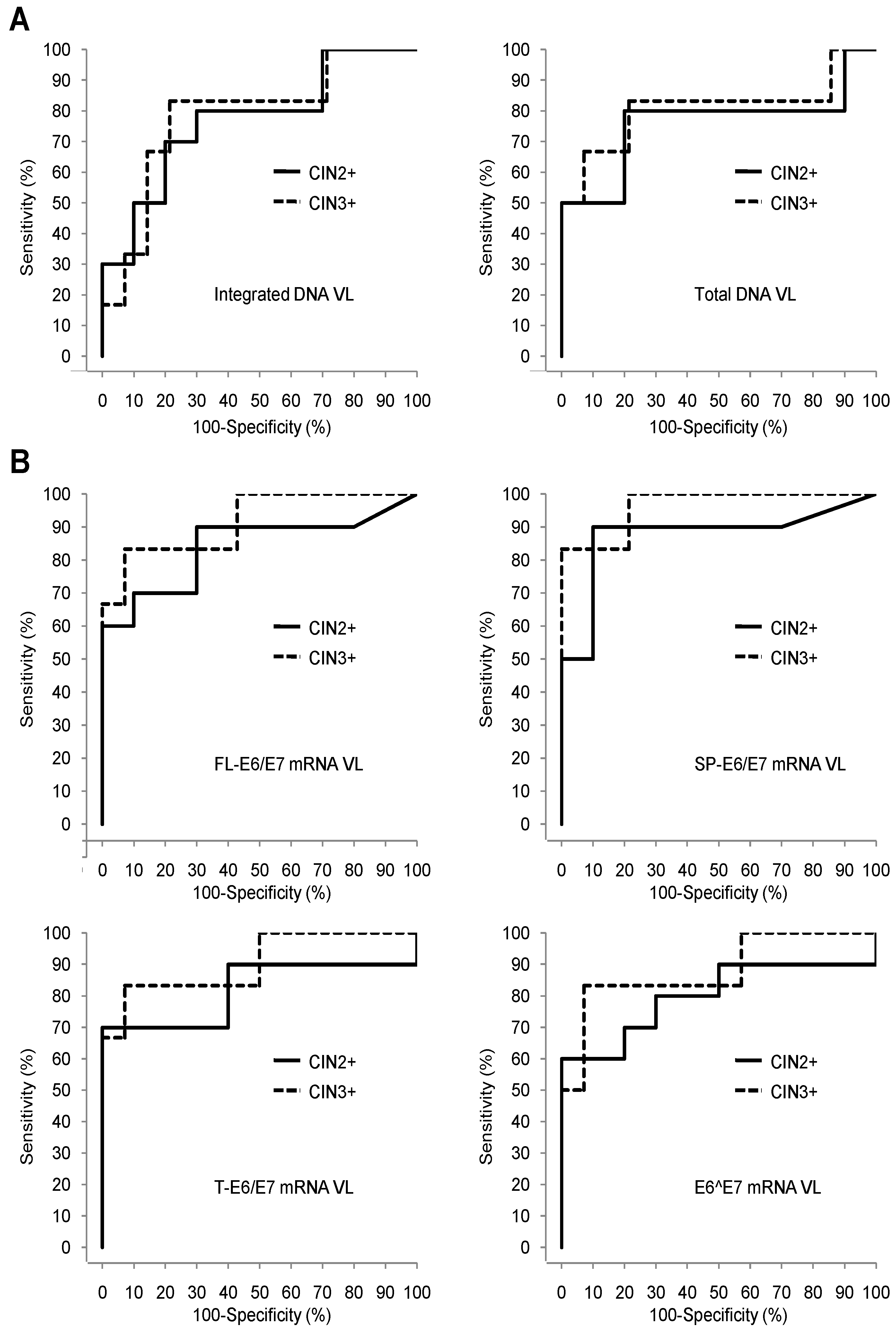
3.4. Comparison of the Pap Test, HPV16 DNA VLs, and HPV16 E6/E7 mRNA VLs Sets Diagnostic Performances for Detection of High Grade Cervical Lesions
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HPVs | human papillomaviruses |
| FL | full length E6/E7 mRNA |
| SP | + spliced E6/E7 mRNA containing intact E7 ORF |
| T | total E6/E7 mRNA corresponding to SP + E6^E7 mRNA |
| VL | viral loads |
| E6^E7 | E6/E7 mRNA containing disrupted E6 and E7 ORFs calculated by the following subtraction T-SP |
| ASC-US | atypical squamous cells of unknown significance |
| LSIL | low-grade squamous intraepithelial lesion |
| CC | cervical cancer |
| CIN | cervical intraepithelial neoplasia |
| CIN2+ | CIN of grade 2 or more: CIN2, CIN3, cancer |
| CIN3+ | CIN of grade 3 or more: CIN3, cancer |
| UCS | uterine cervical smears |
| ASC-H | atypical squamous cells- cannot exclude high grade |
| HSIL | high grade squamous intraepithelial lesion |
| ROC | receiver operating curves |
| AUC | area under roc curve |
| NPV | negative predictive value |
| PPV | positive predictive value |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, 359–386. [Google Scholar] [CrossRef] [PubMed]
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.; Peto, J.; Meijer, C.J.; Munoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Munoz, N. Human papillomavirus and cancer: The epidemiological evidence. J. Clin. Virol. 2000, 19, 1–5. [Google Scholar] [CrossRef]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25, 2–23. [Google Scholar] [CrossRef] [Green Version]
- Bouvard, V.; Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens—Part b: Biological agents. Lancet Oncol. 2009, 10, 321–322. [Google Scholar] [CrossRef]
- De Villiers, E.M.; Fauquet, C.; Broker, T.R.; Bernard, H.U.; zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Munoz, N.; Bosch, F.X.; de Sanjose, S.; Herrero, R.; Castellsague, X.; Shah, K.V.; Snijders, P.J.; Meijer, C.J. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003, 348, 518–527. [Google Scholar] [CrossRef]
- Clifford, G.M.; Smith, J.S.; Plummer, M.; Munoz, N.; Franceschi, S. Human papillomavirus types in invasive cervical cancer worldwide: A meta-analysis. Br. J. Cancer 2003, 88, 63–73. [Google Scholar] [CrossRef]
- de Sanjose, S.; Quint, W.G.; Alemany, L.; Geraets, D.T.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.R.; et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Human papillomaviruses. IARC Monogr. Eval. Carcinog Risks Hum. 2007, 90, 1–636. [Google Scholar]
- French, D.; Lorenzon, L. HPV infections: Basis of neoplastic transformation and related molecular tests. Curr. Pharm. Des. 2013, 19, 1371–1378. [Google Scholar] [PubMed]
- Schlecht, N.F.; Kulaga, S.; Robitaille, J.; Ferreira, S.; Santos, M.; Miyamura, R.A.; Duarte-Franco, E.; Rohan, T.E.; Ferenczy, A.; Villa, L.L.; et al. Persistent human papillomavirus infection as a predictor of cervical intraepithelial neoplasia. JAMA 2001, 286, 3106–3114. [Google Scholar] [CrossRef]
- Comparetto, C.; Borruto, F. Cervical cancer screening: A never-ending developing program. World J. Clin. Cases 2015, 3, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Grosclaude, P. Contributions of the descriptive epidemiology, the registers and the troops. French situation seen by the registers of population. Med. Sci. (Paris) 2007, 23, 22–25. [Google Scholar] [CrossRef] [PubMed]
- SEER Cancer Statistics Review, 1975–2006. Available online: https://seer.cancer.gov/archive/csr/1975_2006/ (accessed on 28 November 2018).
- Cuzick, J.; Arbyn, M.; Sankaranarayanan, R.; Tsu, V.; Ronco, G.; Mayrand, M.H.; Dillner, J.; Meijer, C.J. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine 2008, 26, 29–41. [Google Scholar] [CrossRef]
- Boulanger, J.C.; Fauvet, R.; Urrutiaguer, S.; Drean, Y.; Sevestre, H.; Ganry, O.; Bergeron, C.; Gondry, J. Cytological history of cases of invasive cervical cancer diagnosed in france in 2006. Gynecol. Obstet. Fertil. 2007, 35, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Lorin, L.; Bertaut, A.; Hudry, D.; Beltjens, F.; Roignot, P.; Bone-Lepinoy, M.C.; Douvier, S.; Arveux, P. About invasive cervical cancer: A french population based study between 1998 and 2010. Eur. J. Obstet. Gynecol. Reprod. Biol. 2015, 191, 1–6. [Google Scholar] [CrossRef]
- Wright, T.C., Jr.; Schiffman, M.; Solomon, D.; Cox, J.T.; Garcia, F.; Goldie, S.; Hatch, K.; Noller, K.L.; Roach, N.; Runowicz, C.; et al. Interim guidance for the use of human papillomavirus DNA testing as an adjunct to cervical cytology for screening. Obstet. Gynecol. 2004, 103, 304–309. [Google Scholar] [CrossRef]
- Agorastos, T.; Chatzistamatiou, K.; Katsamagkas, T.; Koliopoulos, G.; Daponte, A.; Constantinidis, T.; Constantinidis, T.C. Primary screening for cervical cancer based on high-risk human papillomavirus (HPV) detection and HPV 16 and HPV 18 genotyping, in comparison to cytology. PLoS ONE 2015, 10, e0119755. [Google Scholar] [CrossRef]
- Ramzan, M.; Noor ul, A.; Ilyas, S.; Umer, M.; Bano, S.; Sarwar, S.; Shahzad, N.; Shakoori, A.R. A cornucopia of screening and diagnostic techniques for human papillomavirus associated cervical carcinomas. J. Virol. Methods 2015, 222, 192–201. [Google Scholar] [CrossRef]
- Tornesello, M.L.; Buonaguro, L.; Giorgi-Rossi, P.; Buonaguro, F.M. Viral and cellular biomarkers in the diagnosis of cervical intraepithelial neoplasia and cancer. Biomed. Res. Int. 2013, 519619. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Chakraborty, C.; Dutta, A.K.; Mandal, R.K.; Roychoudhury, S.; Basu, P.; Panda, C.K. Physical and methylation status of human papillomavirus 16 in asymptomatic cervical infections changes with malignant transformation. J. Clin. Pathol. 2015, 68, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Tsakogiannis, D.; Kyriakopoulou, Z.; Ruether, I.G.; Amoutzias, G.D.; Dimitriou, T.G.; Diamantidou, V.; Kotsovassilis, C.; Markoulatos, P. Determination of human papillomavirus 16 physical status through E1/E6 and E2/E6 ratio analysis. J. Med. Microbiol. 2014, 63, 1716–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; He, Y.F.; Chen, M.; Chen, C.M.; Zhu, Q.J.; Lu, H.; Wei, Z.H.; Li, F.; Zhang, X.X.; Xu, C.J.; et al. Diagnosis of 25 genotypes of human papillomaviruses for their physical statuses in cervical precancerous/cancerous lesions: A comparison of E2/E6E7 ratio-based vs. Multiple E1-L1/E6E7 ratio-based detection techniques. J. Transl. Med. 2014, 12, 282. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Qin, Y.; Yu, L.; Lin, C.; Wang, H.; Cui, J.; Liu, B.; Liao, Y.; Warren, D.; Zhang, X.; et al. Association between human papillomavirus (HPV) 16, HPV18, and other HR-HPV viral load and the histological classification of cervical lesions: Results from a large-scale cross-sectional study. J. Med. Virol. 2017, 89, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Marongiu, L.; Godi, A.; Parry, J.V.; Beddows, S. Human papillomavirus 16, 18, 31 and 45 viral load, integration and methylation status stratified by cervical disease stage. BMC Cancer 2014, 14, 384. [Google Scholar] [CrossRef]
- Origoni, M.; Cristoforoni, P.; Carminati, G.; Stefani, C.; Costa, S.; Sandri, M.T.; Mariani, L.; Preti, M. E6/E7 mrna testing for human papilloma virus-induced high-grade cervical intraepithelial disease (CIN2/CIN3): A promising perspective. Ecancermedicalscience 2015, 9, 533. [Google Scholar] [CrossRef]
- Ajiro, M.; Jia, R.; Zhang, L.; Liu, X.; Zheng, Z.M. Intron definition and a branch site adenosine at nt 385 control RNA splicing of HPV16 E6*I and E7 expression. PLoS ONE 2012, 7, e46412. [Google Scholar] [CrossRef]
- Shirasawa, H.; Jin, M.H.; Shimizu, K.; Akutsu, N.; Shino, Y.; Simizu, B. Transcription-modulatory activity of full-length E6 and E6*I proteins of human papillomavirus type 16. Virology 1994, 203, 36–42. [Google Scholar] [CrossRef]
- Ajiro, M.; Zheng, Z.M. E6^E7, a novel splice isoform protein of human papillomavirus 16, stabilizes viral E6 and E7 oncoproteins via HSP90 and GRP78. Am. Soc. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Chen, J. Signaling pathways in HPV-associated cancers and therapeutic implications. Rev. Med. Virol. 2015, 25, 24–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang-Johanning, F.; Lu, D.W.; Wang, Y.; Johnson, M.R.; Johanning, G.L. Quantitation of human papillomavirus 16 E6 and E7 DNA and rna in residual material from thinprep papanicolaou tests using real-time polymerase chain reaction analysis. Cancer 2002, 94, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.M.; Lee, B.H.; Chang, S.F.; Chien, T.Y.; Huang, S.H.; Yan, C.C.; Cheng, W.F. Type-specific human papillomavirus oncogene messenger RNA levels correlate with the severity of cervical neoplasia. Int. J. Cancer 2010, 127, 622–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, C.C.; Phelps, W.C.; Lindgren, V.; Braun, M.J.; Gonda, M.A.; Howley, P.M. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J. Virol. 1987, 61, 962–971. [Google Scholar] [PubMed]
- Solomon, D.; Davey, D.; Kurman, R.; Moriarty, A.; O’Connor, D.; Prey, M.; Raab, S.; Sherman, M.; Wilbur, D.; Wright, T., Jr.; et al. The 2001 bethesda system: Terminology for reporting results of cervical cytology. JAMA 2002, 287, 2114–2119. [Google Scholar] [CrossRef] [PubMed]
- Peitsaro, P.; Johansson, B.; Syrjanen, S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J. Clin. Microbiol. 2002, 40, 886–891. [Google Scholar] [CrossRef]
- Steinau, M.; Rajeevan, M.S.; Unger, E.R. DNA and RNA references for qRT-PCR assays in exfoliated cervical cells. J. Mol. Diagn. 2006, 8, 113–118. [Google Scholar] [CrossRef]
- Ginsburg, O.; Bray, F.; Coleman, M.P.; Vanderpuye, V.; Eniu, A.; Kotha, S.R.; Sarker, M.; Huong, T.T.; Allemani, C.; Dvaladze, A.; et al. The global burden of women’s cancers: A grand challenge in global health. Lancet 2017, 389, 847–860. [Google Scholar] [CrossRef]
- Mustafa, R.A.; Santesso, N.; Khatib, R.; Mustafa, A.A.; Wiercioch, W.; Kehar, R.; Gandhi, S.; Chen, Y.; Cheung, A.; Hopkins, J.; et al. Systematic reviews and meta-analyses of the accuracy of HPV tests, visual inspection with acetic acid, cytology, and colposcopy. Int. J. Gynaecol. Obstet. 2016, 132, 259–265. [Google Scholar] [CrossRef]
- Burger, E.A.; Kornor, H.; Klemp, M.; Lauvrak, V.; Kristiansen, I.S. HPV mRNA tests for the detection of cervical intraepithelial neoplasia: A systematic review. Gynecol. Oncol. 2011, 120, 430–438. [Google Scholar] [CrossRef]
- Woodman, C.B.; Collins, S.I.; Young, L.S. The natural history of cervical hpv infection: Unresolved issues. Nat. Rev. Cancer 2007, 7, 11–22. [Google Scholar] [CrossRef]
- Ruttkay-Nedecky, B.; Jimenez Jimenez, A.M.; Nejdl, L.; Chudobova, D.; Gumulec, J.; Masarik, M.; Adam, V.; Kizek, R. Relevance of infection with human papillomavirus: The role of the p53 tumor suppressor protein and E6/E7 zinc finger proteins (Review). Int. J. Oncol. 2013, 43, 1754–1762. [Google Scholar] [CrossRef] [PubMed]
- Cricca, M.; Venturoli, S.; Leo, E.; Costa, S.; Musiani, M.; Zerbini, M. Molecular analysis of HPV 16 E6I/E6II spliced mRNAs and correlation with the viral physical state and the grade of the cervical lesion. J. Med. Virol. 2009, 81, 1276–1282. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Dalstein, V.; Waterboer, T.; Clavel, C.; Gissmann, L.; Pawlita, M. The HPV16 transcriptome in cervical lesions of different grades. Mol. Cell. Probes 2011, 25, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Tao, M.; McCoy, J.P., Jr.; Zheng, Z.M. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol. 2006, 80, 4249–4263. [Google Scholar] [CrossRef] [PubMed]
- Pim, D.; Massimi, P.; Banks, L. Alternatively spliced Hpv-18 E6* protein inhibits E6 mediated degradation of p53 and suppresses transformed cell growth. Oncogene 1997, 15, 257–264. [Google Scholar] [CrossRef]

| Primer/Probe Set | Sequences | |
|---|---|---|
| HPV16 E6 | Forward | 5′-GAGAACTGCAATGTTTCAGGACC-3′ |
| Reverse | 5′- TGTATAGTTGTTTGCAGCTCTGTGC-3′ | |
| Probe | JOE-5′-CAGGAGCGACCCAGAAAGTTACCACAGTT-3′-TAMRA | |
| HPV16 E2 | Forward | 5′-AACGAAGTATCCTCTCCTGAAATTATTAG-3′ |
| Reverse | 5′-CCAAGGCGACGGCTTTG-3′ | |
| Probe | FAM-5′-CACCCCGCCGCGACCCATA-3′-TAMRA | |
| β-globin | Forward | 5′-TGCATCTGACTCCTGAGGAGAA-3′ |
| Reverse | 5′-GGGCCTCACCACCAACTTC-3′ | |
| Probe | TET-5′-CTGCCGTTACTGCCCT-3′-TAMRA |
| Primer/Probe Set | Sequences | |
|---|---|---|
| FL-E6/E7 | Forward | 5′-GTGTACTGCAAGCAACAGTTA-3′ |
| Reverse | 5′-CCCATCTCTATATACTATGCATAAATCC-3′ | |
| Probe | FAM-5′-CTGCGACGTGAGGTATATGACTTTGCT-3′-TAMRA | |
| SP-E6/E7 | Forward | 5′-GATTTGCAACCAGAGACAACTG-3′ |
| Reverse | 5′-GCTGGACCATCTATTTCATCCT-3′ | |
| Probe | FAM-5′-TGAGCAATTAAATGACAGCTCAGAGGAGG-3′-TAMRA | |
| T-E6/E7 | Forward | 5′-GACTCTACGCTTCGGTTGTG-3′ |
| Reverse | 5′-TGTGCCCATTAACAGGTCTT-3′ | |
| Probe | FAM-5′-CGTACAAAGCACACACGTAGACATTCG-3′-TAMRA | |
| β-actin | Forward | 5′-GACCCAGATCATGTTTGAGACC-3′ |
| Reverse | 5′-CCAGAGGCGTACAGGGATA-3′ | |
| Probe | FAM-5′-TGTACGTTGCTATCCAGGCTGTGC-3′-TAMRA |
| Viral Load Type | DNA VL (Copies/Cell ± SD (Range)) | E6/E7 mRNA VL (log10 (Copies/106 β-actin mRNA Copies) ±SD (Range)) | |
|---|---|---|---|
| DNA | Total | 0.93 ± 0.2 (0.7–1.06) | |
| Episomal | 0 | ||
| Integrated | 0.93 ± 0.2 (0.7–1.06) | ||
| mRNA | FL-E6/E7 | 4.47 ± 0.18 (4.27–4.59) | |
| SP-E6/E7 | 4.86 ± 0.21 (4.63–5.00) | ||
| T-E6/E7 | 4.93 ± 0.23 (4.68-5.12) | ||
| E6^E7 | 4.09 ± 0.37 (3.73-4.48) |
| Patient ID | Cytology Grade | Histology Grade | HPV16 DNA VL (log10 (Copies/106 Cells)) | HPV16 mRNA VL (log10 (Copies/106 β-actin mRNA Copies)) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Episomal | Integrated | FL-E6/E7 | SP-E6/E7 | T-E6/E7 | E6^E7 | |||
| 31 | Normal | Unlesional | 5.92 | 5.62 | 5.61 | 2.02 | 2.34 | 2.86 | 2.71 |
| 44 | Normal | Unlesional | 6.23 | 5.77 | 6.04 | 0.00 | 0.00 | 2.90 | 2.90 |
| 15 | LSIL | CIN1 | 6.80 | 6.01 | 6.73 | 2.15 | 2.73 | 3.99 | 3.97 |
| 18 | LSIL | CIN1 | 7.56 | 6.86 | 7.46 | 1.78 | 2.15 | 3.50 | 3.48 |
| 34 | Normal | CIN1 | 6.08 | 6.06 | 4.81 | 3.45 | 4.06 | 4.09 | 2.94 |
| 42 | Normal | CIN1 | 5.35 | 5.06 | 5.03 | 1.76 | 0.00 | 3.31 | 3.31 |
| 43 | Normal | CIN1 | 6.59 | 6.35 | 6.22 | 0.00 | 1.59 | 2.95 | 2.93 |
| 45 | ASC-US | CIN1 | 7.61 | 6.76 | 7.55 | 1.80 | 2.49 | 4.34 | 4.34 |
| 51 | LSIL | CIN1 | 6.95 | 6.36 | 6.83 | 2.97 | 0.00 | 4.30 | 4.30 |
| 55 | ASC-US | CIN1 | 5.89 | 5.79 | 5.19 | 2.91 | 3.04 | 3.27 | 2.87 |
| 2 | ASC-H | CIN2 | 6.97 | 6.17 | 6.89 | 2.80 | 3.16 | 3.88 | 3.79 |
| 4 | ASC-US | CIN2 | 5.37 | 4.71 | 5.27 | 0.00 | 0.00 | 1.91 | 1.91 |
| 9 | LSIL | CIN2 | 7.83 | 7.09 | 7.75 | 3.05 | 3.79 | 5.01 | 4.99 |
| 47 | ASC-H | CIN2 | 6.96 | 6.47 | 6.79 | 3.53 | 3.94 | 4.45 | 4.29 |
| 1 | HSIL | CIN3 | 8.09 | 7.48 | 7.97 | 3.52 | 4.19 | 4.72 | 4.56 |
| 5 | ASC-H | CIN3 | 7.19 | 6.55 | 7.08 | 4.25 | 4.81 | 5.10 | 4.79 |
| 41 | HSIL | CIN3 | 5.51 | 5.12 | 5.28 | 2.54 | 3.17 | 3.51 | 3.23 |
| 50 | ASC-H | CIN3 | 7.63 | 6.94 | 7.52 | 4.12 | 5.16 | 5.62 | 5.44 |
| 3 | HSIL | Invasive Cancer | 7.88 | 7.67 | 7.47 | 3.91 | 4.49 | 5.67 | 5.64 |
| 6 | HSIL | Invasive Cancer | 7.97 | 7.62 | 7.72 | 4.11 | 4.81 | 5.26 | 5.07 |
| HPV16 Viral Load Type | CIN2+ Histology Threshold | CIN3+ Histology Threshold | ||||||
|---|---|---|---|---|---|---|---|---|
| Histology <CIN2 | Histology CIN2+ | p-Value (Wilcoxon) | Histology <CIN3 | Histology CIN3+ | p-Value (Wilcoxon) | |||
| DNA VL (log10 (copies/106 cells)) | Total | Mean ± SD | 6.5 ± 0.7 | 7.1 ± 1.0 | 0.0690 | 6.6±0.8 | 7.4 ± 1.0 | 0.0490 |
| Integrated | Mean ± SD | 6.1 ± 1 | 7.0 ± 1.0 | 0.0596 | 6.3±1.0 | 7.2 ± 1.0 | 0.0676 | |
| E6/E7 mRNA VL (log10 (copies/106 β-actin mRNA copies)) | FL-E6/E7 | Mean ± SD | 1.9 ± 1.2 | 3.2 ± 1.3 | 0.0201 | 2.0±1.2 | 3.7 ± 0.6 | 0.0102 |
| SP-E6/E7 | Mean ± SD | 1.8 ± 1.4 | 3.8 ± 1.5 | 0.0112 | 2.1±1.5 | 4.4 ± 0.7 | 0.0048 | |
| T-E6/E7 | Mean ± SD | 3.6 ± 0.6 | 4.5 ± 1.2 | 0.0279 | 3.6±0.8 | 5.0 ± 0.8 | 0.0124 | |
| E6^E7 | Mean ± SD | 3.4 ± 0.6 | 4.4 ± 1.1 | 0.0380 | 3.5±0.8 | 4.8 ± 0.9 | 0.0177 | |
| HPV16 Viral Load Type | CIN2+ Histology Threshold | CIN3+ Histology Threshold | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TV | YI | NPV. % (95%CI) | PPV. % (95%CI) | Se.% (95%CI) | Spe. % (95%CI) | AUC (95%CI) | TV | YI | NPV. % (95%CI) | PPV. % (95%CI) | Se.% (95%CI) | Spe. % (95%CI) | AUC (95%CI) | ||
| DNA | Total | 6.96 | 0.60 | 80 (44–97) | 80 (44–97) | 80 (44–97) | 80 (44–97) | 0.76 (0.52–1.00) | 7.19 | 0.62 | 92 (36–100) | 63 (24–91) | 83 (36–100) | 79 (49–95) | 0.81 (0.53–1.00) |
| Integrated | 6.79 | 0.50 | 78 (40–97) | 73 (39–94) | 80 (44–97) | 70 (35–93) | 0.77 (0.55–0.99) | 7.08 | 0.62 | 92 (62–100) | 63 (24–91) | 83 (36–100) | 79 (49–95) | 0.79 (0.54–1.00) | |
| mRNA | FL-E6/E7 | 2.54 | 0.60 | 88 (47–100) | 75 (43–95) | 90 (56–100) | 70 (35–99) | 0.84 (0.65–1.00) | 2.54 | 0.57 | 100 (63–100) | 50 (21–79) | 100 (54–100) | 57 (29–82) | 0.92 (0.77–1.00) |
| SP-E6/E7 | 3.16 | 0.80 | 90 (56–100) | 90 (56–100) | 90 (56–100) | 90 (56–100) | 0.88 (0.69–1.00) | 3.17 | 0.79 | 100 (72–100) | 67 (30–93) | 100 (54–100) | 79 (49–95) | 0.97 (0.89–1.00) | |
| T-E6/E7 | 3.51 | 0.50 | 86 (42–100) | 69 (39–91) | 90 (56–100) | 60 (26–88) | 0.82 (0.60–1.00) | 4.72 | 0.76 | 93 (66–100) | 83 (36–100) | 83 (36–100) | 93 (66–100) | 0.91 (0.73–1.00) | |
| E6^E7 | 3.23 | 0.40 | 83 (36–100) | 64 (35–87) | 90 (56–100) | 50 (19–81) | 0.80 (0.58–1.00) | 3.23 | 0.43 | 100 (54–100) | 43 (18–71) | 100 (54–100) | 43 (18–71) | 0.88 (0.69–1.00) | |
| Pap test | NA | NA | 83 (52–98) | 100 (63–100) | 80 (44–97) | 100 (69–100) | NA | NA | NA | 100 (74–100) | 75 (35–97) | 100 (54–100) | 86 (57–98) | NA | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camus, C.; Vitale, S.; Loubatier, C.; Pénaranda, G.; Khiri, H.; Plauzolles, A.; Carcopino, X.; Halfon, P.; Giordanengo, V. Quantification of HPV16 E6/E7 mRNA Spliced Isoforms Viral Load as a Novel Diagnostic Tool for Improving Cervical Cancer Screening. J. Clin. Med. 2018, 7, 530. https://doi.org/10.3390/jcm7120530
Camus C, Vitale S, Loubatier C, Pénaranda G, Khiri H, Plauzolles A, Carcopino X, Halfon P, Giordanengo V. Quantification of HPV16 E6/E7 mRNA Spliced Isoforms Viral Load as a Novel Diagnostic Tool for Improving Cervical Cancer Screening. Journal of Clinical Medicine. 2018; 7(12):530. https://doi.org/10.3390/jcm7120530
Chicago/Turabian StyleCamus, Claire, Sébastien Vitale, Céline Loubatier, Guillaume Pénaranda, Hacène Khiri, Anne Plauzolles, Xavier Carcopino, Philippe Halfon, and Valérie Giordanengo. 2018. "Quantification of HPV16 E6/E7 mRNA Spliced Isoforms Viral Load as a Novel Diagnostic Tool for Improving Cervical Cancer Screening" Journal of Clinical Medicine 7, no. 12: 530. https://doi.org/10.3390/jcm7120530





