Differentiating Malignant from Benign Pigmented or Non-Pigmented Skin Tumours—A Pilot Study on 3D Hyperspectral Imaging of Complex Skin Surfaces and Convolutional Neural Networks
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Clinical Study
2.2. Patient Demographics and Lesion Characteristics
2.3. The SICSURFIS Hyperspectral Imager
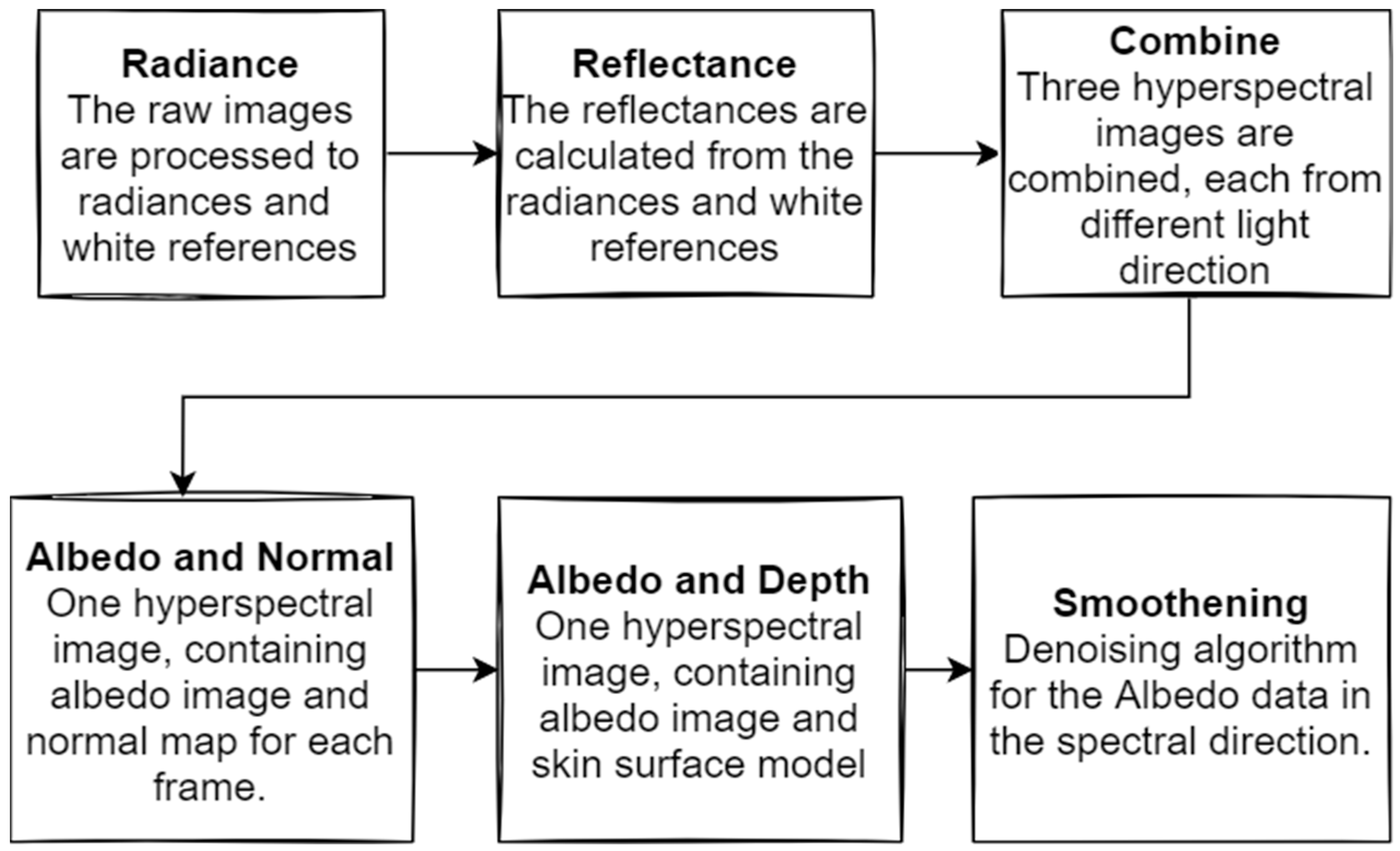
2.4. Data Pre-Processing
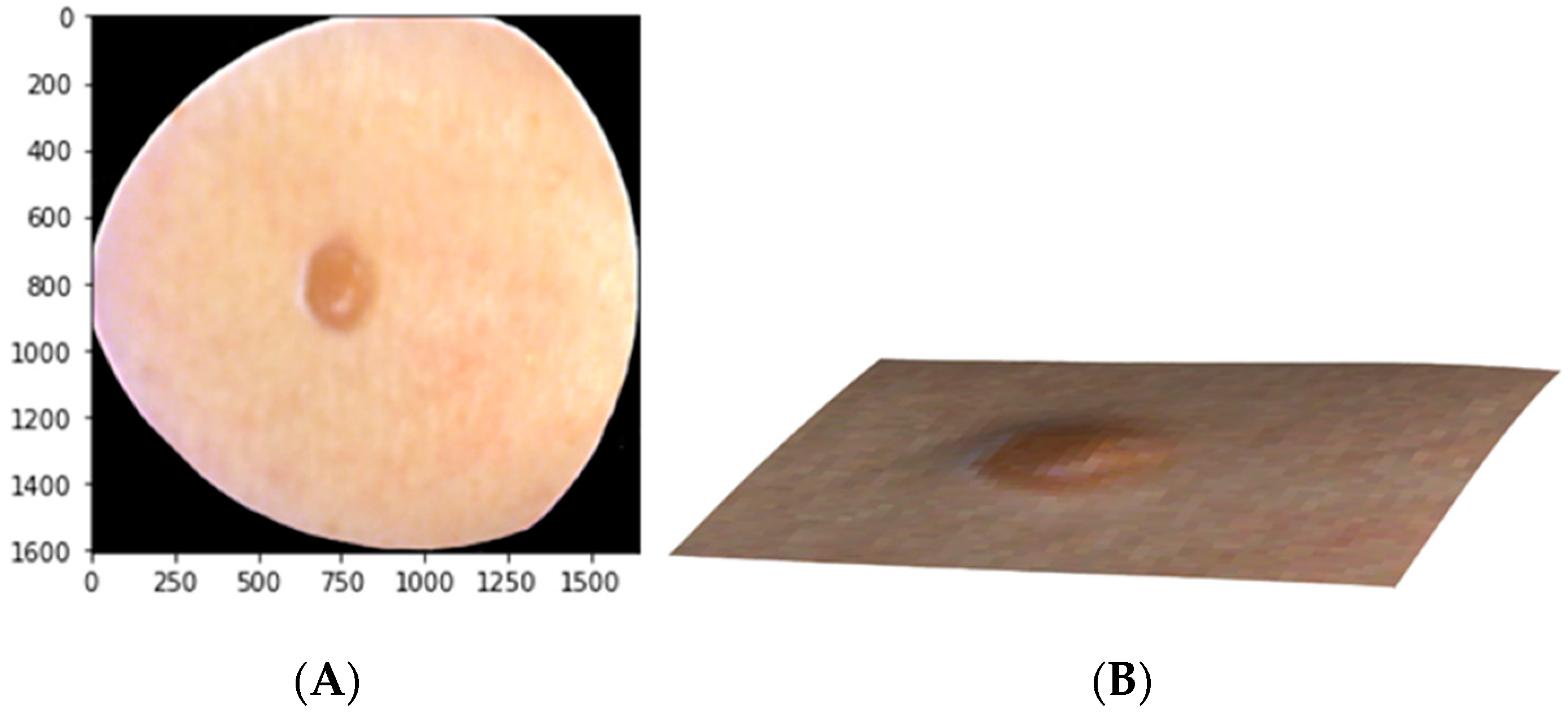
2.5. Raw Data Pre-Processing
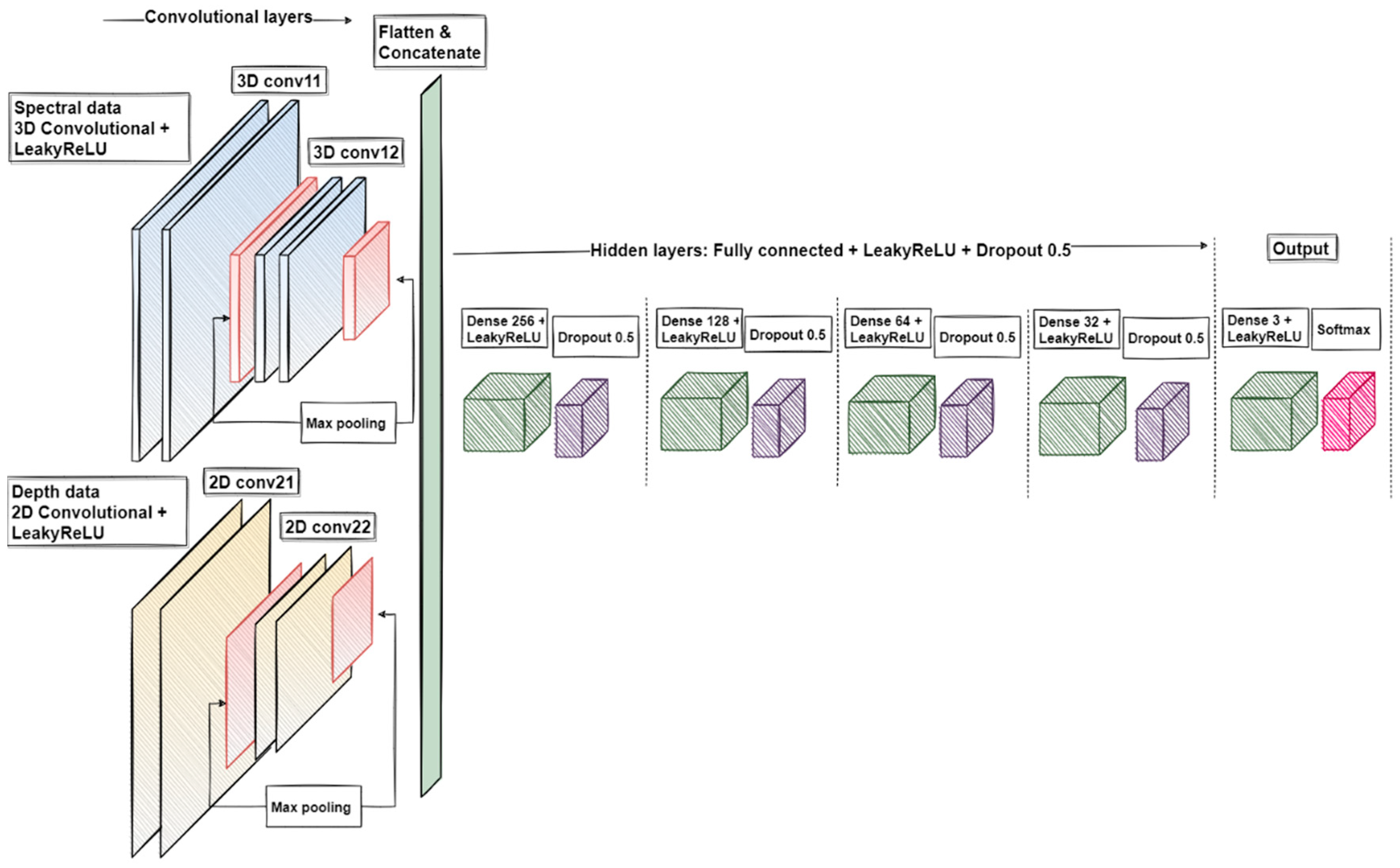
2.6. Machine Learning Pre-Processing
2.7. Data Analysis
3. Results
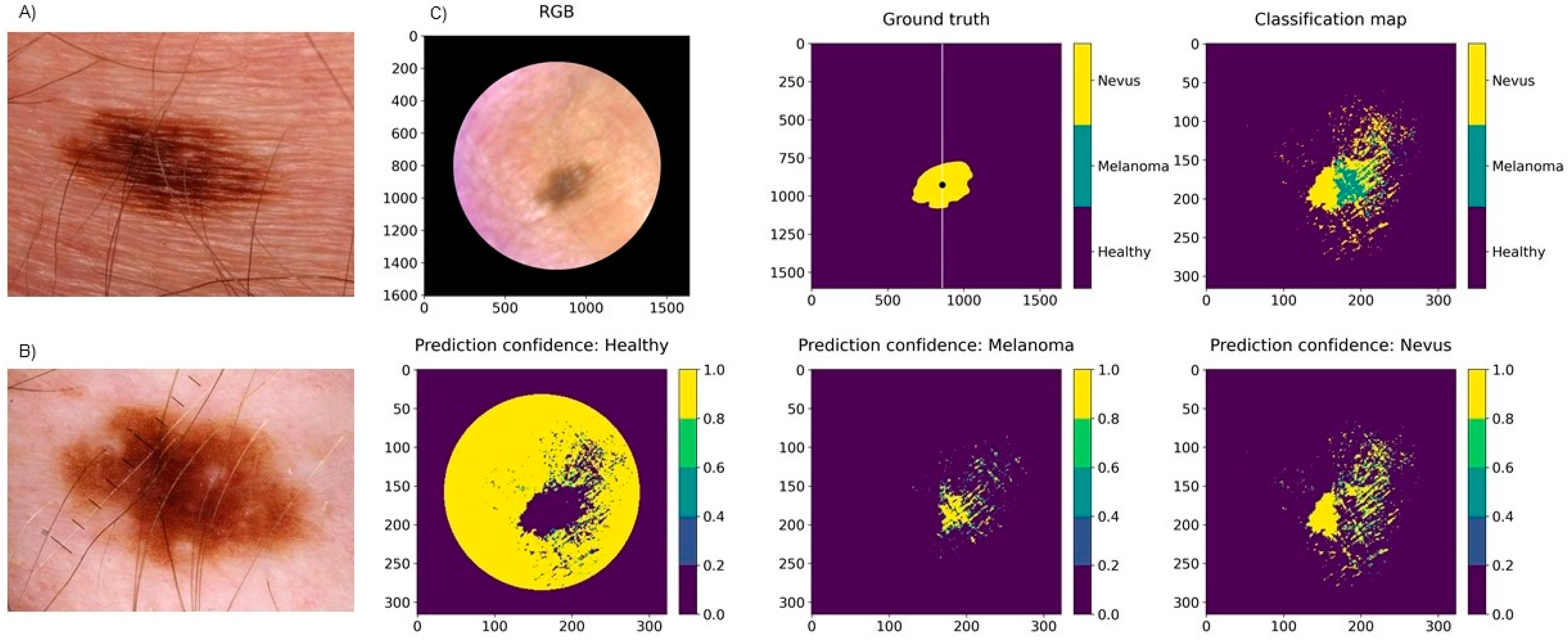
3.1. Classification Results for Pigmented Lesions
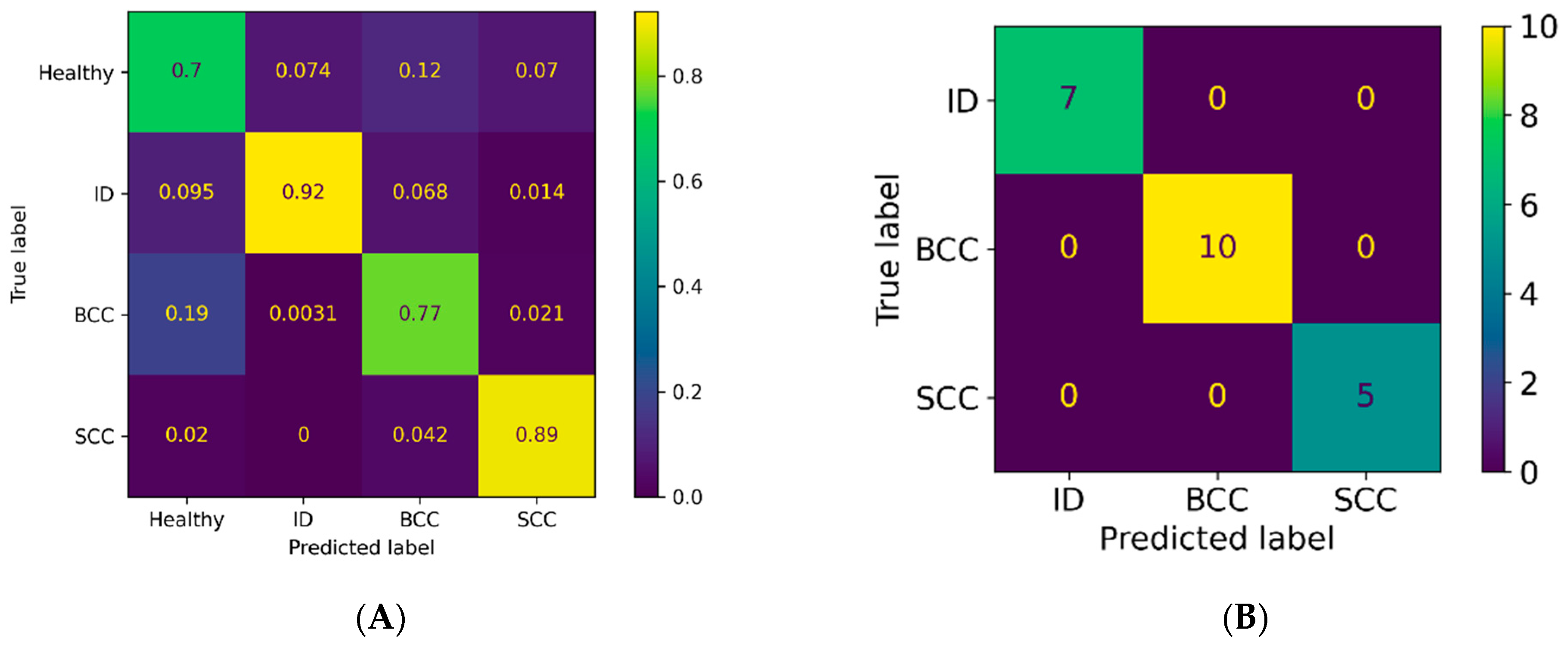
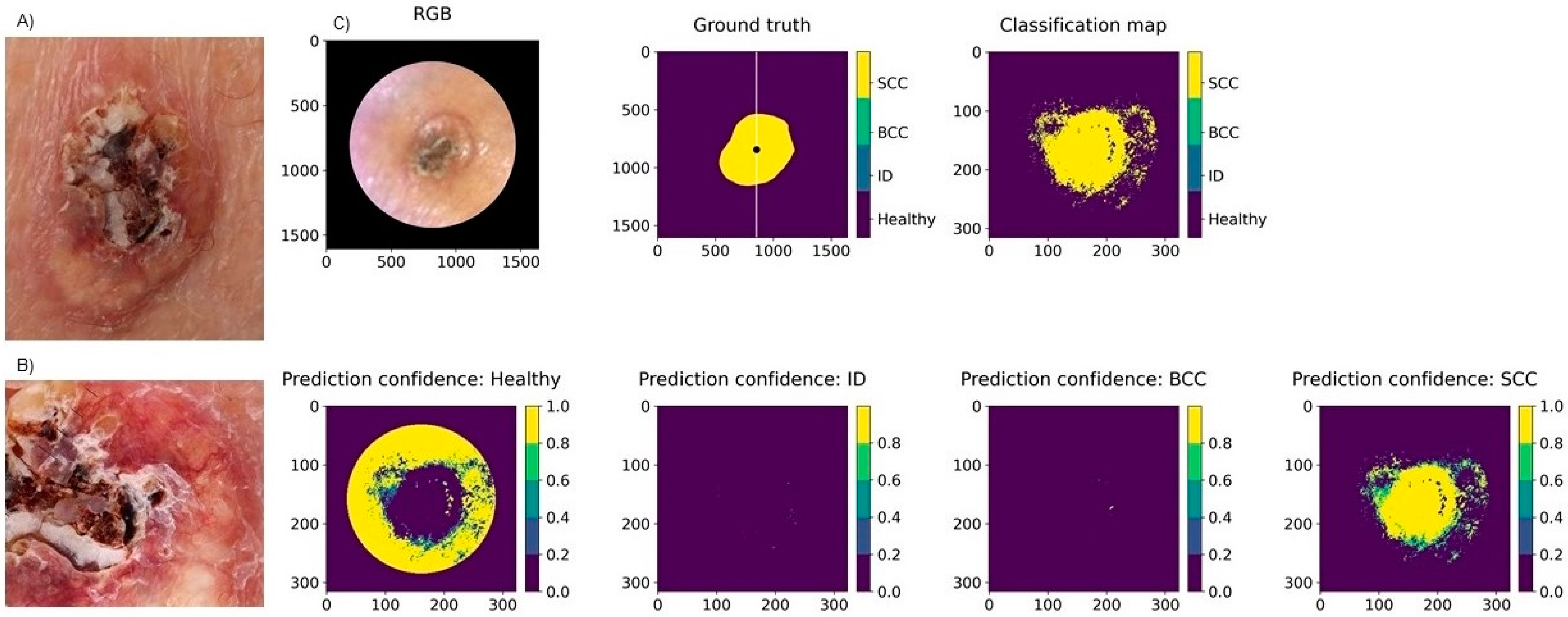
3.2. Classification Results for Non-Pigmented Lesions
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organisation. Radiation: Ultraviolet (UV) Radiation and Skin Cancer. Available online: https://www.who.int/uv/faq/skincancer/en/index1.html (accessed on 10 January 2022).
- Yélamos, O.; Braun, R.P.; Liopyris, K.; Wolner, Z.J.; Kerl, K.; Gerami, P.; Marghoob, A.A. Usefulness of Dermoscopy to Improve the Clinical and Histopathologic Diagnosis of Skin Cancers. J. Am. Acad. Dermatol. 2019, 80, 365–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapey, J.; Xie, Y.; Nabavi, E.; Bradford, R.; Saeed, S.R.; Ourselin, S.; Vercauteren, T. Intraoperative Multispectral and Hyperspectral Label-free Imaging: A Systematic Review of in Vivo Clinical Studies. J. Biophotonics 2019, 12, e201800455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, G.; Fei, B. Medical Hyperspectral Imaging: A Review. J. Biomed. Opt. 2014, 19, 010901. [Google Scholar] [CrossRef] [PubMed]
- Rey-Barroso, L.; Peña-Gutiérrez, S.; Yáñez, C.; Burgos-Fernández, F.J.; Vilaseca, M.; Royo, S. Optical Technologies for the Improvement of Skin Cancer Diagnosis: A Review. Sensors 2021, 21, 252. [Google Scholar] [CrossRef] [PubMed]
- Neittaanmäki-Perttu, N.; Grönroos, M.; Tani, T.; Pölönen, I.; Ranki, A.; Saksela, O.; Snellman, E. Detecting Field Cancerization Using a Hyperspectral Imaging System. Lasers Surg. Med. 2013, 45, 410–417. [Google Scholar] [CrossRef]
- Neittaanmäki-Perttu, N.; Karppinen, T.; Tani, T.; Snellman, E.; Grönroos, M. Long-Term Outcome of Low-Concentration Hexyl-5-Aminolaevulinate Daylight Photodynamic Therapy for Treatment of Actinic Keratoses. Acta Derm. Venerol. 2017, 97, 120–121. [Google Scholar] [CrossRef]
- Räsänen, J.; Salmivuori, M.; Pölönen, I.; Grönroos, M.; Neittaanmäki, N. Hyperspectral Imaging Reveals Spectral Differences and Can Distinguish Malignant Melanoma from Pigmented Basal Cell Carcinomas: A Pilot Study. Acta Derm. Venereol. 2021, 101, adv00405. [Google Scholar] [CrossRef]
- Neittaanmäki-Perttu, N.; Grönroos, M.; Jeskanen, L.; Pölönen, I.; Ranki, A.; Saksela, O.; Snellman, E. Delineating Margins of Lentigo Maligna Using a Hyperspectral Imaging System. Acta Derm. Venereol. 2015, 95, 549–552. [Google Scholar] [CrossRef] [Green Version]
- Salmivuori, M.; Neittaanmäki, N.; Pölönen, I.; Jeskanen, L.; Snellman, E.; Grönroos, M. Hyperspectral Imaging System in the Delineation of Ill-defined Basal Cell Carcinomas: A Pilot Study. J. Eur. Acad. Dermatol. 2019, 33, 71–78. [Google Scholar] [CrossRef]
- Christensen, G.B.; Nagaoka, T.; Kiyohara, Y.; Johansson, I.; Ingvar, C.; Nakamura, A.; Sota, T.; Nielsen, K. Clinical Performance of a Novel Hyperspectral Imaging Device for Cutaneous Melanoma and Pigmented Skin Lesions in Caucasian Skin. Skin Res. Technol. 2021, 27, 803–809. [Google Scholar] [CrossRef]
- Hirano, G.; Nemoto, M.; Kimura, Y.; Kiyohara, Y.; Koga, H.; Yamazaki, N.; Christensen, G.; Ingvar, C.; Nielsen, K.; Nakamura, A.; et al. Automatic Diagnosis of Melanoma Using Hyperspectral Data and GoogLeNet. Skin Res. Technol. 2020, 26, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Nemoto, M.; Kimura, Y.; Kiyohara, Y.; Koga, H.; Yamazaki, N.; Christensen, G.; Ingvar, C.; Nielsen, K.; Nakamura, A.; et al. Performance Improvement of Automated Melanoma Diagnosis System by Data Augmentation. ABE 2020, 9, 62–70. [Google Scholar] [CrossRef] [Green Version]
- Leon, R.; Martinez-Vega, B.; Fabelo, H.; Ortega, S.; Melian, V.; Castaño, I.; Carretero, G.; Almeida, P.; Garcia, A.; Quevedo, E.; et al. Non-Invasive Skin Cancer Diagnosis Using Hyperspectral Imaging for In-Situ Clinical Support. J. Clin. Med. 2020, 9, 1662. [Google Scholar] [CrossRef] [PubMed]
- Pölönen, I.; Rahkonen, S.; Annala, L.; Neittaanmäki, N. Convolutional neural networks in cancer detection using spatial and spectral domain. Proc. SPIE 2019, 2019, 108510B. [Google Scholar] [CrossRef] [Green Version]
- Abend, K.; Harley, T.; Chandrasekaran, B.; Hughes, G. Comments on “On the Mean Accuracy of Statistical Pattern Recognizers” by Hughes, G.F. IEEE Trans. Inform. Theory 1969, 15, 420–423. [Google Scholar] [CrossRef]
- Ding, S.; Keal, C.A.; Zhao, L.; Yu, D. Dimensionality Reduction and Classification for Hyperspectral Image Based on Robust Supervised ISOMAP. J. Int. Prod. Eng. 2022, 39, 19–29. [Google Scholar] [CrossRef]
- Raita-Hakola, A.M.; Annala, L.; Lindholm, V.; Trops, R.; Näsilä, A.; Saari, H.; Ranki, A.; Pölönen, I. FPI based Hyperspectral Imager for the Complex Surfaces-Calibration, Illumination and Applications. Sensors 2022. submitted. [Google Scholar]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against Machine: Diagnostic Performance of a Deep Learning Convolutional Neural Network for Dermoscopic Melanoma Recognition in Comparison to 58 Dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Ferrante di Ruffano, L.; Takwoingi, Y.; Dinnes, J.; Chuchu, N.; Bayliss, S.E.; Davenport, C.; Matin, R.N.; Godfrey, K.; O’Sullivan, C.; Gulati, A.; et al. Computer-Assisted Diagnosis Techniques (Dermoscopy and Spectroscopy-Based) for Diagnosing Skin Cancer in Adults. Cochrane Database Syst. Rev. 2018, 2018, CD013186. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Akay, B.N.; Argenziano, G.; Blum, A.; Braun, R.P.; Cabo, H.; Gourhant, J.-Y.; Kreusch, J.; Lallas, A.; et al. Expert-Level Diagnosis of Nonpigmented Skin Cancer by Combined Convolutional Neural Networks. JAMA Dermatol. 2019, 155, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Codella, N.; Akay, B.N.; Argenziano, G.; Braun, R.P.; Cabo, H.; Gutman, D.; Halpern, A.; Helba, B.; Hofmann-Wellenhof, R.; et al. Comparison of the Accuracy of Human Readers versus Machine-Learning Algorithms for Pigmented Skin Lesion Classification: An Open, Web-Based, International, Diagnostic Study. Lancet Oncol. 2019, 20, 938–947. [Google Scholar] [CrossRef]
- Trops, R.; Hakola, A.-M.; Jääskeläinen, S.; Näsilä, A.; Annala, L.; Eskelinen, M.A.; Saari, H.; Pölönen, I.; Rissanen, A. Miniature MOEMS Hyperspectral Imager with Versatile Analysis Tools. In Proceedings of the MOEMS and Miniaturized Systems XVIII, San Francisco, CA, USA, 2–7 February 2019; Piyawattanametha, W., Park, Y.-H., Zappe, H., Eds.; SPIE: Washington, DC, USA, 2019; p. 31. [Google Scholar]
- University of Jyväskylä, Spectral Imaging Laboratory. Available online: https://www.jyu.fi/it/en/research/our-laboratories/hsi (accessed on 10 January 2022).
- Barun, V.V.; Ivanov, A.P.; Volotovskaya, A.V.; Ulashchik, V.S. Absorption Spectra and Light Penetration Depth of Normal and Pathologically Altered Human Skin. J. Appl. Spectrosc. 2007, 74, 430–439. [Google Scholar] [CrossRef]
- van der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T. Scikit-Image: Image Processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [Green Version]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. Tensor Flow Large-Scale Machine Learning on Heterogeneous Systems. Available online: https://arxiv.org/abs/1603.04467 (accessed on 10 January 2022).
- Nagaoka, T.; Kiyohara, Y.; Koga, H.; Nakamura, A.; Saida, T.; Sota, T. Modification of a Melanoma Discrimination Index Derived from Hyperspectral Data: A Clinical Trial Conducted in 2 Centers between March 2011 and December 2013. Skin Res. Technol. 2015, 21, 278–283. [Google Scholar] [CrossRef]
- Pardo, A.; Gutiérrez-Gutiérrez, J.A.; Lihacova, I.; López-Higuera, J.M.; Conde, O.M. On the Spectral Signature of Melanoma: A Non-Parametric Classification Framework for Cancer Detection in Hyperspectral Imaging of Melanocytic Lesions. Biomed. Opt. Express 2018, 9, 6283. [Google Scholar] [CrossRef] [Green Version]
- Heibel, H.D.; Hooey, L.; Cockerell, C.J. A Review of Noninvasive Techniques for Skin Cancer Detection in Dermatology. Am. J. Clin. Dermatol. 2020, 21, 513–524. [Google Scholar] [CrossRef]
- Pezzini, C.; Kaleci, S.; Chester, J.; Farnetani, F.; Longo, C.; Pellacani, G. Reflectance Confocal Microscopy Diagnostic Accuracy for Malignant Melanoma in Different Clinical Settings: Systematic Review and Meta-analysis. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 2268–2279. [Google Scholar] [CrossRef]
- Wessels, R.; de Bruin, D.M.; Relyveld, G.N.; Faber, D.J.; Vincent, A.D.; Sanders, J.; van Leeuwen, T.G.; Ruers, T.J.M. Functional Optical Coherence Tomography of Pigmented Lesions. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Gambichler, T.; Schmid-Wendtner, M.H.; Plura, I.; Kampilafkos, P.; Stücker, M.; Berking, C.; Maier, T. A Multicentre Pilot Study Investigating High-Definition Optical Coherence Tomography in the Differentiation of Cutaneous Melanoma and Melanocytic Naevi. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Dinnes, J.; Deeks, J.J.; Chuchu, N.; Matin, R.N.; Wong, K.Y.; Aldridge, R.B.; Durack, A.; Gulati, A.; Chan, S.A.; Johnston, L.; et al. Visual Inspection and Dermoscopy, Alone or in Combination, for Diagnosing Keratinocyte Skin Cancers in Adults. Cochrane Database Syst. Rev. 2018, 2018, CD011901. [Google Scholar] [CrossRef] [PubMed]
- Dinnes, J.; Deeks, J.J.; Chuchu, N.; Saleh, D.; Bayliss, S.E.; Takwoingi, Y.; Davenport, C.; Patel, L.; Matin, R.N.; O’Sullivan, C.; et al. Reflectance Confocal Microscopy for Diagnosing Keratinocyte Skin Cancers in Adults. Cochrane Database Syst. Rev. 2018, 2018, CD013191. [Google Scholar] [CrossRef]
- Ferrante di Ruffano, L.; Dinnes, J.; Deeks, J.J.; Chuchu, N.; Bayliss, S.E.; Davenport, C.; Takwoingi, Y.; Godfrey, K.; O’Sullivan, C.; Matin, R.N.; et al. Optical Coherence Tomography for Diagnosing Skin Cancer in Adults. Cochrane Database Syst. Rev. 2018, 2018, CD013189. [Google Scholar] [CrossRef]
- Terstappen, K.; Suurküla, M.; Hallberg, H.; Ericson, M.B.; Wennberg, A.-M. Poor Correlation between Spectrophotometric Intracutaneous Analysis and Histopathology in Melanoma and Nonmelanoma Lesions. J. Biomed. Opt. 2013, 18, 061223. [Google Scholar] [CrossRef] [Green Version]
- Rajaram, N.; Reichenberg, J.S.; Migden, M.R.; Nguyen, T.H.; Tunnell, J.W. Pilot Clinical Study for Quantitative Spectral Diagnosis of Non-Melanoma Skin Cancer: Spectral Diagnosis of Skin Cancers. Lasers Surg. Med. 2010, 42, 876–887. [Google Scholar] [CrossRef] [Green Version]
- Neittaanmäki, N.; Salmivuori, M.; Pölönen, I.; Jeskanen, L.; Ranki, A.; Saksela, O.; Snellman, E.; Grönroos, M. Hyperspectral Imaging in Detecting Dermal Invasion in Lentigo Maligna Melanoma. Br. J. Dermatol. 2017, 177, 1742–1744. [Google Scholar] [CrossRef]



| PATIENTS | 33 | LESIONS | 42 |
| Mean age | 68 | Mean diameter | 10.3 mm (2–30 mm) |
| Males | 16 (48%) | Diagnosis | |
| Females | 17 (52%) | Pigmented lesions: | |
| Fitzpatrick skin type | MM | 7 (17%) | |
| I | 9 (27%) | Superficial spreading MM | 7 (17%) |
| II | 12 (36%) | PN | 13 (31%) |
| III | 12 (36%) | Junctional nevi | 5 (12%) |
| History of skin cancer | 17 (52%) | Compound nevi | 8 (19%) |
| BCC | 8 (24%) | High-grade dysplastic PN | 2 (5%) |
| MM | 7 (21%) | Low-grade dysplastic PN | 3 (7%) |
| SCC | 2 (6%) | Nevus recurrence | 1 (2%) |
| BCC + MM | 3 (9%) | Non-pigmented lesions: | |
| History of other cancers | 6 (18%) | BCC | 10 (24%) |
| Breast | 3 (9%) | Nodular | 6 (14%) |
| GI | 2 (6%) | Nodular + superficial | 3 (7%) |
| Prostate | 2 (6%) | Superficial | 1 (2%) |
| Blood | 1 (3%) | SCC | 5 (12%) |
| Immunosuppression | 4 (12%) | ID | 7 (17%) |
| Radiation therapy | 3 (9%) | Location: | |
| Multiple nevus syndrome | 2 (6%) | Head/neck | 13 (31%) |
| Dysplastic nevi | 6 (18%) | Torso | 21 (50%) |
| Family history of skin cancer | 4 (12%) | Upper extremities | 2 (5%) |
| Multiple nevus syndrome in the family | 4 (12%) | Lower extremities | 6 (14%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lindholm, V.; Raita-Hakola, A.-M.; Annala, L.; Salmivuori, M.; Jeskanen, L.; Saari, H.; Koskenmies, S.; Pitkänen, S.; Pölönen, I.; Isoherranen, K.; et al. Differentiating Malignant from Benign Pigmented or Non-Pigmented Skin Tumours—A Pilot Study on 3D Hyperspectral Imaging of Complex Skin Surfaces and Convolutional Neural Networks. J. Clin. Med. 2022, 11, 1914. https://doi.org/10.3390/jcm11071914
Lindholm V, Raita-Hakola A-M, Annala L, Salmivuori M, Jeskanen L, Saari H, Koskenmies S, Pitkänen S, Pölönen I, Isoherranen K, et al. Differentiating Malignant from Benign Pigmented or Non-Pigmented Skin Tumours—A Pilot Study on 3D Hyperspectral Imaging of Complex Skin Surfaces and Convolutional Neural Networks. Journal of Clinical Medicine. 2022; 11(7):1914. https://doi.org/10.3390/jcm11071914
Chicago/Turabian StyleLindholm, Vivian, Anna-Maria Raita-Hakola, Leevi Annala, Mari Salmivuori, Leila Jeskanen, Heikki Saari, Sari Koskenmies, Sari Pitkänen, Ilkka Pölönen, Kirsi Isoherranen, and et al. 2022. "Differentiating Malignant from Benign Pigmented or Non-Pigmented Skin Tumours—A Pilot Study on 3D Hyperspectral Imaging of Complex Skin Surfaces and Convolutional Neural Networks" Journal of Clinical Medicine 11, no. 7: 1914. https://doi.org/10.3390/jcm11071914






