Correlation of KRAS G12C Mutation and High PD-L1 Expression with Clinical Outcome in NSCLC Patients Treated with Anti-PD1 Immunotherapy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Mutational Analyses in Tissue Samples
2.2.1. Next-Generation Sequencing Analyses
2.2.2. Fluorescence In Situ Hybridization
2.2.3. Immunohistochemistry
2.3. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Elkrief, A.; Joubert, P.; Florescu, M.; Tehfe, M.; Blais, N.; Routy, B. Therapeutic landscape of metastatic non-small-cell lung cancer in Canada in 2020. Curr. Oncol. 2020, 27, 52–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanna, N.H.; Robinson, A.G.; Temin, S.; Baker, S., Jr.; Brahmer, J.R.; Ellis, P.M.; Gaspar, L.E.; Haddad, R.Y.; Hesketh, P.J.; Jain, D.; et al. Therapy for Stage IV Non-Small-Cell Lung Cancer With Driver Alterations: ASCO and OH (CCO) Joint Guideline Update. J. Clin. Oncol. 2021, 39, 1040–1091. [Google Scholar] [CrossRef] [PubMed]
- Hanna, N.H.; Schneider, B.J.; Temin, S.; Baker, S., Jr.; Brahmer, J.; Ellis, P.M.; Gaspar, L.E.; Haddad, R.Y.; Hesketh, P.J.; Jain, D.; et al. Therapy for Stage IV Non-Small-Cell Lung Cancer Without Driver Alterations: ASCO and OH (CCO) Joint Guideline Update. J. Clin. Oncol. 2020, 38, 1608–1632. [Google Scholar] [CrossRef]
- Azzoli, C.G.; Baker, S., Jr.; Temin, S.; Pao, W.; Aliff, T.; Brahmer, J.; Johnson, D.H.; Laskin, J.L.; Masters, G.; Milton, D.; et al. American Society of Clinical Oncology Clinical Practice Guideline update on chemotherapy for stage IV non-small-cell lung cancer. J. Clin. Oncol. 2009, 27, 6251–6266. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Updated Analysis of KEYNOTE-024: Pembrolizumab Versus Platinum-Based Chemotherapy for Advanced Non-Small-Cell Lung Cancer With PD-L1 Tumor Proportion Score of 50% or Greater. J. Clin. Oncol. 2019, 37, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, L.; Rodríguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Giaccone, G.; de Marinis, F.; Reinmuth, N.; Vergnenegre, A.; Barrios, C.H.; Morise, M.; Felip, E.; Andric, Z.; Geater, S.; et al. Atezolizumab for First-Line Treatment of PD-L1-Selected Patients with NSCLC. N. Engl. J. Med. 2020, 383, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, L.; Luft, A.; Vicente, D.; Tafreshi, A.; Gümüş, M.; Mazières, J.; Hermes, B.; Çay Şenler, F.; Csőszi, T.; Fülöp, A.; et al. Pembrolizumab plus Chemotherapy for Squamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379, 2040–2051. [Google Scholar] [CrossRef] [PubMed]
- Horn, L.; Mansfield, A.S.; Szczęsna, A.; Havel, L.; Krzakowski, M.; Hochmair, M.J.; Huemer, F.; Losonczy, G.; Johnson, M.L.; Nishio, M.; et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379, 2220–2229. [Google Scholar] [CrossRef] [PubMed]
- Planchard, D.; Popat, S.; Kerr, K.; Novello, S.; Smit, E.F.; Faivre-Finn, C.; Mok, T.S.; Reck, M.; Van Schil, P.E.; Hellmann, M.D.; et al. Metastatic Non-Small Cell Lung Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Updated version published 15 September 2020 by the ESMO Guidelines Committee. Available online: https://www.esmo.org/content/download/347819/6934778/1/ESMO-CPG-mNSCLC-15SEPT2020.pdf (accessed on 20 August 2021).
- Frigola, J.; Navarro, A.; Carbonell, C.; Callejo, A.; Iranzo, P.; Cedrés, S.; Martinez-Marti, A.; Pardo, N.; Saoudi-Gonzalez, N.; Martinez, D.; et al. Molecular profiling of long-term responders to immune checkpoint inhibitors in advanced non-small cell lung cancer. Mol. Oncol. 2021, 15, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Bruno, D.; Dowlati, A. Immunotherapy in EGFR mutant non-small cell lung cancer: When, who and how? Transl. Lung Cancer Res. 2019, 8, 710–714. [Google Scholar] [CrossRef] [PubMed]
- Calles, A.; Riess, J.W.; Brahmer, J.R. Checkpoint Blockade in Lung Cancer With Driver Mutation: Choose the Road Wisely. Am. Soc. Clin. Oncol. Educ. Book 2020, 40, 372–384. [Google Scholar] [CrossRef]
- Roeper, J.; Kurz, S.; Grohé, C.; Griesinger, F. Optimizing therapy sequence to prevent patient attrition in EGFR mutation-positive advanced or metastatic NSCLC. Future Oncol. 2021, 17, 471–486. [Google Scholar] [CrossRef]
- Nadler, E.; Arondekar, B.; Aguilar, K.M.; Zhou, J.; Chang, J.; Zhang, X.; Pawar, V. Treatment patterns and clinical outcomes in patients with advanced non-small cell lung cancer initiating first-line treatment in the US community oncology setting: A real-world retrospective observational study. J. Cancer Res. Clin. Oncol. 2021, 147, 671–690. [Google Scholar] [CrossRef]
- Abernethy, A.P.; Arunachalam, A.; Burke, T.; McKay, C.; Cao, X.; Sorg, R.; Carbone, D.P. Real-world first-line treatment and overall survival in non-small cell lung cancer without known EGFR mutations or ALK rearrangements in US community oncology setting. PLoS ONE 2017, 12, e0178420. [Google Scholar] [CrossRef] [PubMed]
- Guisier, F.; Dubos-Arvis, C.; Viñas, F.; Doubre, H.; Ricordel, C.; Ropert, S.; Janicot, H.; Bernardi, M.; Fournel, P.; Lamy, R.; et al. Efficacy and Safety of Anti-PD-1 Immunotherapy in Patients With Advanced NSCLC With BRAF, HER2, or MET Mutations or RET Translocation: GFPC 01-2018. J. Thorac. Oncol. 2020, 15, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Hsu, M.; Cohen, R.B.; Langer, C.J.; Mamtani, R.; Aggarwal, C. Association Between KRAS Variant Status and Outcomes With First-line Immune Checkpoint Inhibitor-Based Therapy in Patients With Advanced Non-Small-Cell Lung Cancer. JAMA Oncol. 2021, 7, 937–939. [Google Scholar] [CrossRef]
- Lee, C.K.; Man, J.; Lord, S.; Cooper, W.; Links, M.; Gebski, V.; Herbst, R.S.; Gralla, R.J.; Mok, T.; Yang, J.C. Clinical and Molecular Characteristics Associated With Survival Among Patients Treated With Checkpoint Inhibitors for Advanced Non-Small Cell Lung Carcinoma: A Systematic Review and Meta-analysis. JAMA Oncol. 2018, 4, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Y.; Zhong, W.Z.; Zhang, X.C.; Su, J.; Xie, Z.; Liu, S.Y.; Tu, H.Y.; Chen, H.J.; Sun, Y.L.; Zhou, Q.; et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2017, 23, 3012–3024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biton, J.; Mansuet-Lupo, A.; Pécuchet, N.; Alifano, M.; Ouakrim, H.; Arrondeau, J.; Boudou-Rouquette, P.; Goldwasser, F.; Leroy, K.; Goc, J.; et al. TP53, STK11, and EGFR Mutations Predict Tumor Immune Profile and the Response to Anti-PD-1 in Lung Adenocarcinoma. Clin. Cancer Res. 2018, 24, 5710–5723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Zeng, D.; Ou, Q.; Liu, S.; Li, A.; Chen, Y.; Lin, D.; Gao, Q.; Zhou, H.; Liao, W.; et al. Association of Survival and Immune-Related Biomarkers with Immunotherapy in Patients with Non-Small Cell Lung Cancer: A Meta-analysis and Individual Patient-Level Analysis. JAMA Netw. Open 2019, 2, e196879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbour, K.C.; Jordan, E.; Kim, H.R.; Dienstag, J.; Yu, H.A.; Sanchez-Vega, F.; Lito, P.; Berger, M.; Solit, D.B.; Hellmann, M.; et al. Effects of Co-occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non-Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 334–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skoulidis, F.; Li, B.T.; Dy, G.K.; Price, T.J.; Falchook, G.S.; Wolf, J.; Italiano, A.; Schuler, M.; Borghaei, H.; Barlesi, F.; et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N. Engl. J. Med. 2021, 384, 2371–2381. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.; Bernasconi, B.; Merlo, E.; Balzarini, P.; Vermi, W.; Riva, A.; Chiaravalli, A.M.; Frattini, M.; Sahnane, N.; Facchetti, F.; et al. ALK testing in lung adenocarcinoma: Technical aspects to improve FISH evaluation in daily practice. J. Thorac. Oncol. 2015, 10, 595–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuta, K.; Kohno, T.; Yoshida, A.; Shimada, Y.; Asamura, H.; Furuta, K.; Kushima, R. RET-rearranged non-small-cell lung carcinoma: A clinicopathological and molecular analysis. Br. J. Cancer 2014, 110, 1571–1578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bubendorf, L.; Büttner, R.; Al-Dayel, F.; Dietel, M.; Elmberger, G.; Kerr, K.; López-Ríos, F.; Marchetti, A.; Öz, B.; Pauwels, P.; et al. Testing for ROS1 in non-small cell lung cancer: A review with recommendations. Virchows Arch. 2016, 469, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef]
- Grambsch, P.; Therneau, T. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika 1994, 81, 515–526. [Google Scholar] [CrossRef]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 2 March 2022).
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 2 March 2022).
- Wickham, H.; Bryan, J. readxl: Read Excel Files. R Package Version 1.3.1. 2019. Available online: https://CRAN.R-project.org/package=readxl (accessed on 2 March 2022).
- Therneau, T. A Package for Survival Analysis in R. R Package Version 3.2-7. 2020. Available online: https://CRAN.R-project.org/package=survival (accessed on 2 March 2022).
- Wickham, H.; François, R.; Henry, L.; Müller, K. dplyr: A Grammar of Data Manipulation. R Package Version 1.0.2. 2020. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 2 March 2022).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Tang, Y.; Horikoshi, M.; Li, W. ggfortify: Unified Interface to Visualize Statistical Result of Popular R Packages. R J. 2016, 8, 478–489. [Google Scholar] [CrossRef] [Green Version]
- Kassambara, A.; Kosinski, M.; Biecek, P. survminer: Drawing Survival Curves Using ‘ggplot2’. R Package Version 0.4.8. 2020. Available online: https://CRAN.R-project.org/package=survminer (accessed on 2 March 2022).
- Fisher, R. The Logic of Inductive Inference. J. R. Stat. Soc. 1935, 98, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Greenland, S.; Senn, S.J.; Rothman, K.J.; Carlin, J.B.; Poole, C.; Goodman, S.N.; Altman, D.G. Statistical tests, P values, confidence intervals, and power: A guide to misinterpretations. Eur. J. Epidemiol. 2016, 31, 337–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hastings, K.; Yu, H.A.; Wei, W.; Sanchez-Vega, F.; DeVeaux, M.; Choi, J.; Rizvi, H.; Lisberg, A.; Truini, A.; Lydon, C.A.; et al. EGFR mutation subtypes and response to immune checkpoint blockade treatment in non-small-cell lung cancer. Ann. Oncol. 2019, 30, 1311–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Factor (%) | Entire Sample | G12C+ | G12C− | Statistical Test between Groups to Assess Balance |
|---|---|---|---|---|
| N | 44 | 11 (25%) | 33 (75%) | NA |
| Number of Females | 19 (43%) | 4 (21%) | 15 (79%) | Fisher’s Exact p = 0.73 |
| Presence of Brain Metastases | 11 (25%) | 2 (18%) | 9 (27%) | Fisher’s Exact p = 0.70 |
| Smokers, actual or former | 42 (95%) | 11 (100%) | 31 (94%) | Fisher’s Exact p = 1 |
| Median Age | 69 | 71 | 68 | H(1) = 1.43, p = 0.23 |
| Median ECOG | 1 | 1 | 1 | H(1) = 0.68, p = 0.68 |
| Group | Progression Free Survival | Overall Survival | ||||
|---|---|---|---|---|---|---|
| Events | Median Time to Event (Days) | 95% CI (Days) | Events | Median Time to Event (Days) | 95% CI (Days) | |
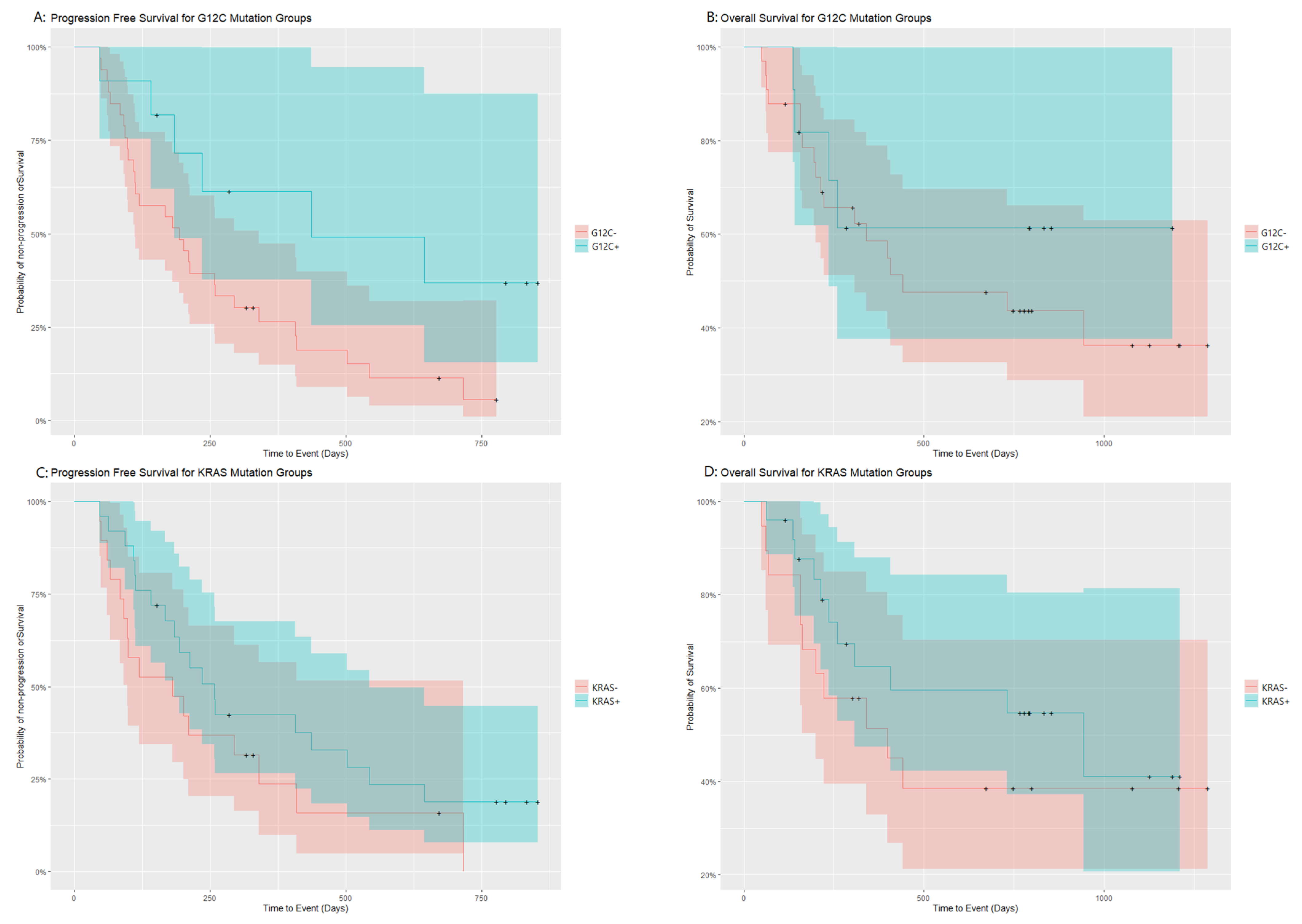
| Entire Population | 35 | 211 | 168–407 | 22 | 731 | 307-NA |
| KRAS+ | 19 | 258 | 98-NA | 11 | 944 | 307-NA |
| KRAS− | 16 | 181 | 184–544 | 11 | 398 | 200-NA |
| Log rank test | χ2(1) = 1.8 p = 0.18 | χ2(1) = 0.9 p = 0.35 | ||||
| G12C+ | 6 | 437 | 184-NA | 4 | NA | 236-NA |
| G12C− | 29 | 194 | 112–340 | 18 | 442 | 307-NA |
| Log rank test | χ2(1) = 4.5 p = 0.03 | χ2(1) = 0.5 p = 0.47 | ||||
| Risk Factor | Model 1 | Model 2 | ||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | p | Hazard Ratio | 95% CI | p | |
| G12C+ | 0.42 | 0.16–1.08 | 0.07 | 0.27 | 0.10–0.76 | 0.01 |
| ECOG | 1.01 | 0.68–1.50 | 0.95 | 1.14 | 0.68–1.92 | 0.61 |
| Past/current Smoker | 0.63 | 0.14–2.85 | 0.55 | 0.62 | 0.14–2.85 | 0.54 |
| Presence of brain metastases | 1.12 | 0.63–1.97 | 0.70 | 1.08 | 0.63–1.86 | 0.78 |
| Likelihood ratio test | χ2(4) = 5.49 | 0.2 | χ2(4) = 10.73 | 0.03 | ||
| Wald Test | χ2(4) = 4.74 | 0.3 | χ2(4) = 8.44 | 0.08 | ||
| Log Rank Test | χ2(4) = 5.07 | 0.2 | χ2(4) = 9.58 | 0.05 | ||
| Concordance | 0.59 | 0.65 | ||||
| Risk Factor | Model 1 | Model 2 | ||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | p | Hazard Ratio | 95% CI | p | |
| KRAS+ | 0.60 | 0.30–1.18 | 0.14 | 0.46 | 0.22–0.95 | 0.04 |
| ECOG | 1.16 | 0.78–1.71 | 0.46 | 1.04 | 0.68–1.58 | 0.85 |
| Past/current Smoker | 0.43 | 0.10–1.88 | 0.26 | 0.39 | 0.09–1.72 | 0.21 |
| Presence of brain metastases | 1.37 | 0.79–2.36 | 0.26 | 1.61 | 0.93–2.80 | 0.09 |
| Likelihood ratio test | χ2(4) = 3.98 | 0.4 | χ2(4) = 6.64 | 0.2 | ||
| Wald Test | χ2(4) = 4.1 | 0.4 | χ2(4) = 6.8 | 0.1 | ||
| Log Rank Test | χ2(4) = 4.17 | 0.4 | χ2(4) = 7.1 | 0.1 | ||
| Concordance | 0.59 | 0.62 | ||||
| Group | PFS Events | |
|---|---|---|
| No Event | Event | |
| KRAS G12C+ | 5 | 6 |
| Other KRAS mut | 1 | 13 |
| Fisher’s Exact p | 0.03 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cefalì, M.; Epistolio, S.; Ramelli, G.; Mangan, D.; Molinari, F.; Martin, V.; Freguia, S.; Mazzucchelli, L.; Froesch, P.; Frattini, M.; et al. Correlation of KRAS G12C Mutation and High PD-L1 Expression with Clinical Outcome in NSCLC Patients Treated with Anti-PD1 Immunotherapy. J. Clin. Med. 2022, 11, 1627. https://doi.org/10.3390/jcm11061627
Cefalì M, Epistolio S, Ramelli G, Mangan D, Molinari F, Martin V, Freguia S, Mazzucchelli L, Froesch P, Frattini M, et al. Correlation of KRAS G12C Mutation and High PD-L1 Expression with Clinical Outcome in NSCLC Patients Treated with Anti-PD1 Immunotherapy. Journal of Clinical Medicine. 2022; 11(6):1627. https://doi.org/10.3390/jcm11061627
Chicago/Turabian StyleCefalì, Marco, Samantha Epistolio, Giulia Ramelli, Dylan Mangan, Francesca Molinari, Vittoria Martin, Stefania Freguia, Luca Mazzucchelli, Patrizia Froesch, Milo Frattini, and et al. 2022. "Correlation of KRAS G12C Mutation and High PD-L1 Expression with Clinical Outcome in NSCLC Patients Treated with Anti-PD1 Immunotherapy" Journal of Clinical Medicine 11, no. 6: 1627. https://doi.org/10.3390/jcm11061627






