Is Brain Network Efficiency Reduced in Young Survivors of Acute Lymphoblastic Leukemia?—Evidence from Individual-Based Morphological Brain Network Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Clinical Assessment
2.3. MRI Data Acquisition
2.4. MRI Data Preprocessing
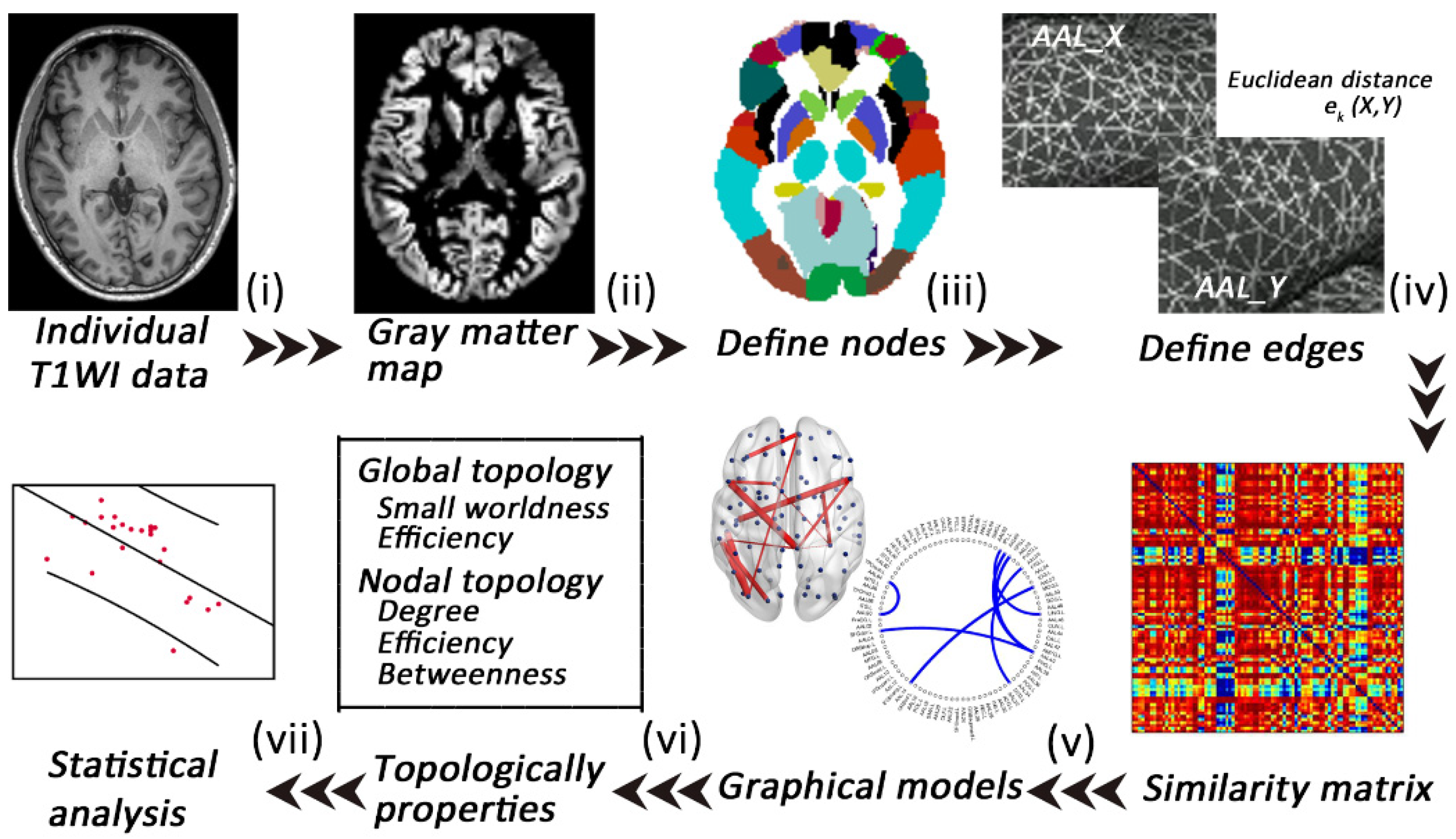
2.5. Extraction of Individual Morphological Brain Networks
2.6. Analyses of Network Properties
2.7. Statistical Analysis
2.7.1. Demographic and Clinical Variable Statistics
2.7.2. Network Topological Metric Statistics
2.7.3. Network-Based Statistics Analysis
2.7.4. Correlation Analysis
3. Results
3.1. Demographic and Clinical Characteristics
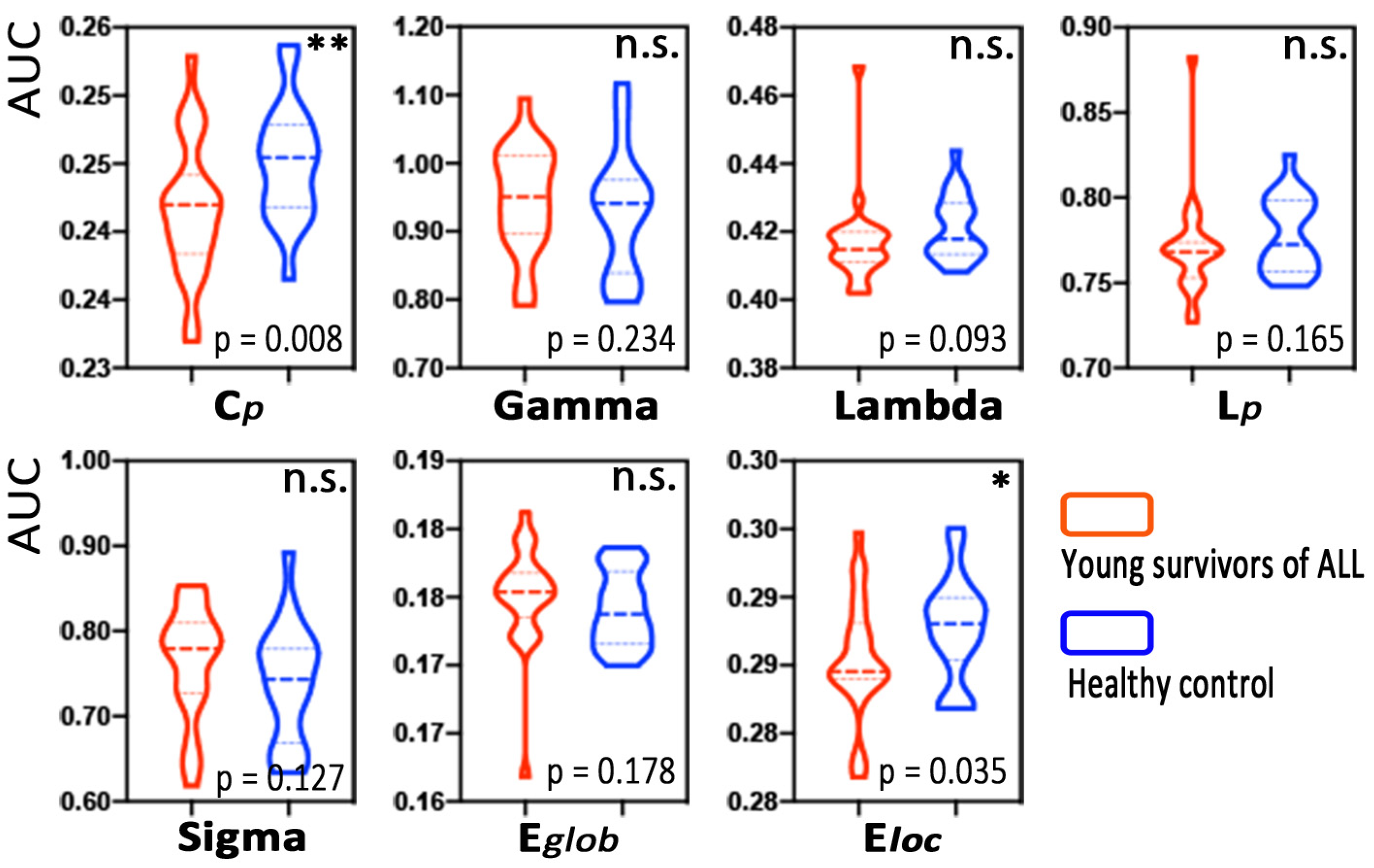
3.2. Alterations in Global Morphological Network Properties
3.3. Alterations in Nodal Morphological Network Properties
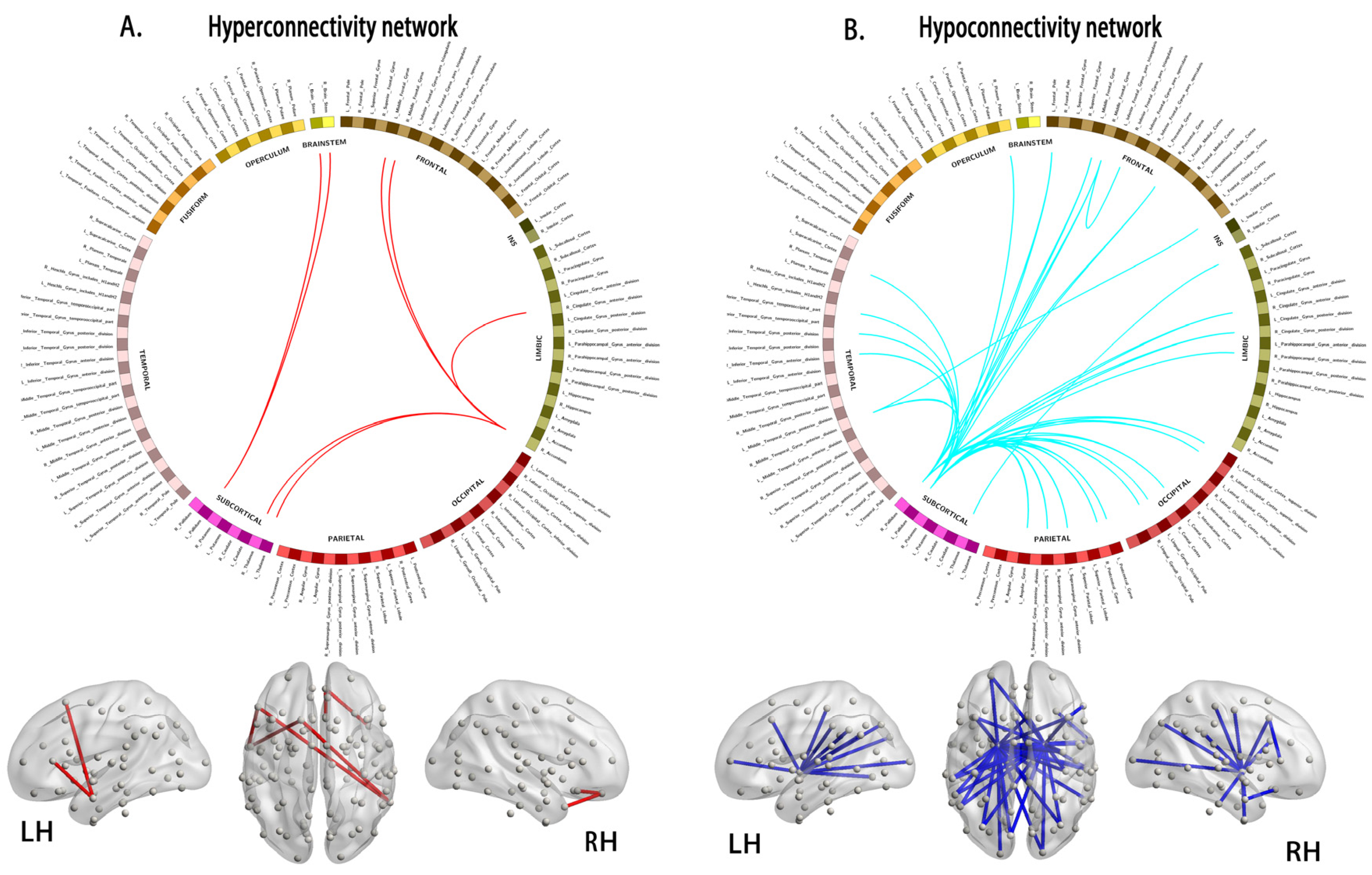
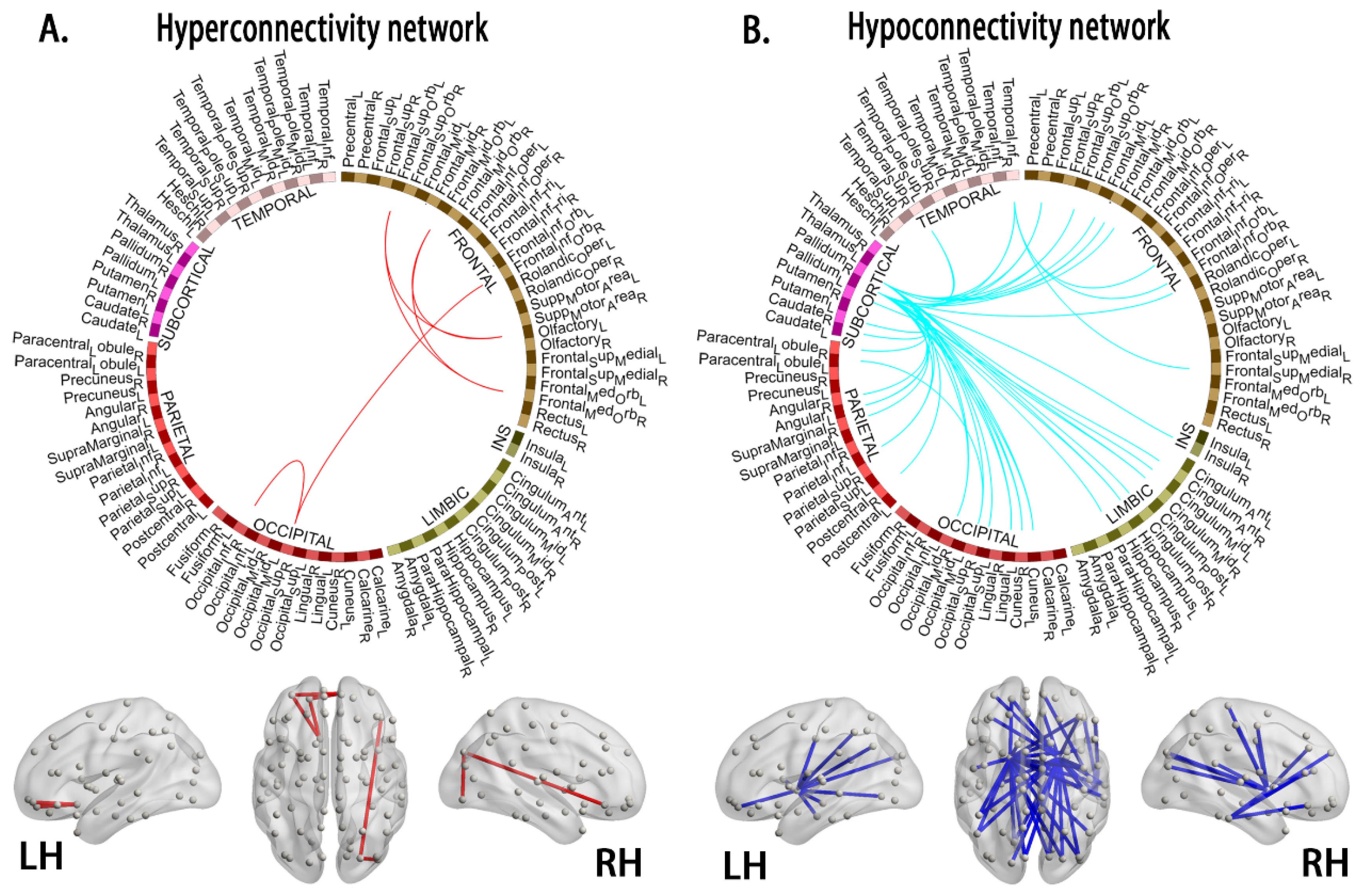
3.4. Alterations in Connections Based on NBS Analyses
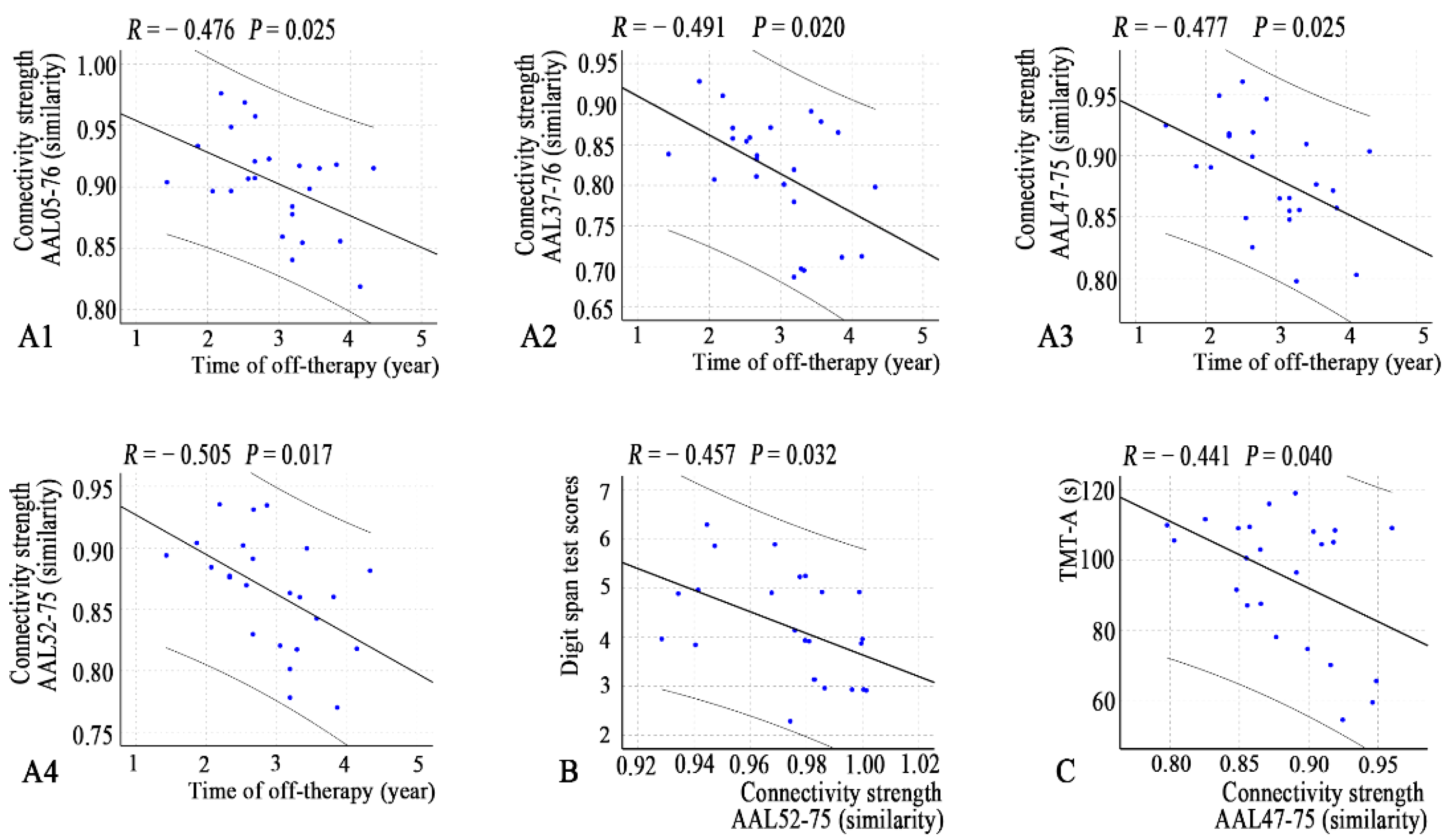
3.5. Relationships between Network Properties and Clinical Variables
4. Discussion
4.1. Decreased Segregation at the Global Level
4.2. Alterations in Nodal Centrality in Young Survivors of ALL
4.3. Hyperconnectivity and Hypoconnectivity in Young Survivors of ALL
4.4. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Brain Regions | AAL Label | Brain Network # | p Values | ||
|---|---|---|---|---|---|
| Nodal Degree | Nodal Betweenness | Nodal Efficiency | |||
| Young survivors of ALL > HC | |||||
| Left inferior frontal gyrus, Triang (IFGtriang) | 13 | Frontal-parietal network | 0.0560 | 0.0322 * | 0.2391 |
| Right Rolandic operculum (ROL) | 18 | Auditory network | 0.0458 * | 0.4599 | 0.0210 * |
| Right supplementary motor area (SMA) | 20 | Cingulo-opercular network | 0.0148 * | 0.4613 | 0.0120 * |
| Left olfactory cortex (OLF) | 21 | Subcortical network | 0.0022 ** | 0.0052 ** | 0.0006 *** |
| Right olfactory cortex | 22 | Subcortical network | 0.0332 * | 0.0914 | 0.0068 ** |
| Right superior frontal gyrus, medial orbital (ORBsupmed) | 26 | Default mode network | 0.0104 * | 0.1702 | 0.0124 * |
| Right posterior cingulum gyrus (PCG) | 36 | Default mode network | 0.0199 * | 0.0468 * | 0.0286 * |
| Right superior occipital gyrus (SOG) | 50 | Visual network | 0.0360 * | 0.0293 * | 0.0622 |
| Right supramarginal gyrus (SMG) | 64 | Auditory network | 0.4265 | 0.0140 * | 0.4369 |
| Left angular gyrus (ANG) | 65 | Default mode network | 0.3893 | 0.0299 * | 0.4693 |
| Right Heschl’s gyrus (HES) | 80 | Auditory network | 0.0762 | 0.0456 * | 0.0702 |
| Young survivors of ALL < HC | |||||
| Left inferior frontal gyrus, opercular part (IFGoperc) | 11 | Salience network | 0.1292 | 0.1765 | 0.0202 * |
| Left hippocampus (HIP) | 37 | - | 0.0370 * | 0.0810 | 0.0379 * |
| Left middle occipital gyrus (MOG) | 51 | Visual network | 0.0760 | 0.3769 | 0.0378 * |
| Right MOG | 52 | Visual network | 0.0324 * | 0.2255 | 0.0184 * |
| Right fusiform gyrus (FFG) | 56 | - | 0.2635 | 0.0090 ** | 0.4815 |
| Left inferior parietal lobe (IPL) | 61 | Frontal-parietal network | 0.0174 * | 0.0418 * | 0.0372 * |
| Left lenticular nucleus, pallidum (PAL) | 75 | Subcortical network | 0.0004 *** | 0.0002 *** | 0.0004 *** |
| Right PAL | 76 | Subcortical network | 0.0130 * | 0.0076 ** | 0.0194 * |
| Left temporal pole: superior temporal gyrus (TPOsup) | 83 | Default mode network | 0.0350 * | 0.4725 | 0.0358 * |
| Right inferior temporal gyrus (ITG) | 90 | - | 0.0462 * | 0.2585 | 0.0472 * |
| Node 1 | Node 2 | t-Values | ||
|---|---|---|---|---|
| AAL | Labels Regions (Abbr.) | AAL | Labels Regions (Abbr.) | |
| Hyperconnectivity morphological subnetworks | ||||
| 5 | Frontal_Sup_Orb_L | 21 | Olfactory_L | 3.402 |
| 5 | Frontal_Sup_Orb_L | 26 | Frontal_Med_Orb_R | 3.767 |
| 9 | Frontal_Mid_Orb_L | 21 | Olfactory_L | 3.526 |
| 9 | Frontal_Mid_Orb_L | 26 | Frontal_Med_Orb_R | 3.139 |
| 16 | Frontal_Inf_Orb_R | 50 | Occipital_Sup_R | 4.047 |
| 50 | Occipital_Sup_R | 54 | Occipital_Inf_R | 2.982 |
| Hypoconnectivity morphological subnetworks | ||||
| 2 | Precentral_R | 75 | Pallidum_L | −3.471 |
| 2 | Precentral_R | 76 | Pallidum_R | −3.965 |
| 5 | Frontal_Sup_Orb_L | 75 | Pallidum_L | −3.135 |
| 5 | Frontal_Sup_Orb_L | 76 | Pallidum_R | −3.386 |
| 7 | Frontal_Mid_L | 76 | Pallidum_R | −2.956 |
| 8 | Frontal_Mid_R | 75 | Pallidum_L | −3.191 |
| 8 | Frontal_Mid_R | 76 | Pallidum_R | −3.054 |
| 9 | Frontal_Mid_Orb_L | 76 | Pallidum_R | −3.054 |
| 14 | Frontal_Inf_Tri_R | 90 | Temporal_Inf_R | −2.976 |
| 16 | Frontal_Inf_Orb_R | 90 | Temporal_Inf_R | −3.629 |
| 17 | Rolandic_Oper_L | 75 | Pallidum_L | −3.017 |
| 24 | Frontal_Sup_Medial_R | 90 | Temporal_Inf_R | −3.467 |
| 30 | Insula_R | 75 | Pallidum_L | −3.269 |
| 32 | Cingulum_Ant_R | 76 | Pallidum_R | −3.022 |
| 33 | Cingulum_Mid_L | 75 | Pallidum_L | −3.227 |
| 34 | Cingulum_Mid_R | 75 | Pallidum_R | −3.590 |
| 34 | Cingulum_Mid_R | 76 | Pallidum_R | −3.293 |
| 36 | Cingulum_Post_R | 76 | Pallidum_R | −3.059 |
| 37 | Hippocampus_L | 75 | Pallidum_L | −3.201 |
| 37 | Hippocampus_L | 76 | Pallidum_R | −2.984 |
| 39 | ParaHippocampal_L | 75 | Pallidum_L | −3.238 |
| 45 | Cuneus_L | 75 | Pallidum_L | −3.319 |
| 45 | Cuneus_L | 76 | Pallidum_R | −3.417 |
| 46 | Cuneus_R | 75 | Pallidum_L | −3.846 |
| 46 | Cuneus_R | 76 | Pallidum_R | −3.642 |
| 47 | Lingual_L | 75 | Pallidum_L | −3.878 |
| 47 | Lingual_L | 76 | Pallidum_R | −3.375 |
| 49 | Occipital_Sup_L | 76 | Pallidum_R | −3.355 |
| 50 | Occipital_Sup_R | 69 | Paracentral_Lobule_L | −3.039 |
| 50 | Occipital_Sup_R | 76 | Pallidum_R | −3.886 |
| 52 | Occipital_Mid_R | 75 | Pallidum_L | −3.410 |
| 52 | Occipital_Mid_R | 76 | Pallidum_R | −3.295 |
| 58 | Postcentral_R | 76 | Pallidum_R | −3.108 |
| 61 | Parietal_Inf_L | 75 | Pallidum_L | −3.323 |
| 61 | Parietal_Inf_L | 76 | Pallidum_R | −3.047 |
| 64 | SupraMarginal_R | 76 | Pallidum_R | −2.924 |
| 65 | Angular_L | 75 | Pallidum_L | −4.112 |
| 65 | Angular_L | 76 | Pallidum_R | −3.616 |
| 66 | Angular_R | 75 | Pallidum_L | −3.014 |
| 70 | Paracentral_Lobule_R | 76 | Pallidum_R | −3.266 |
| 71 | Caudate_L | 75 | Pallidum_L | −3.064 |
| 72 | Caudate_R | 75 | Pallidum_L | −2.966 |
| 74 | Putamen_R | 75 | Pallidum_L | −3.288 |
| 75 | Pallidum_L | 82 | Temporal_Sup_R | −3.356 |
| 75 | Pallidum_L | 90 | Temporal_Inf_R | −3.665 |
| 76 | Pallidum_R | 82 | Temporal_Sup_R | −3.059 |
| 76 | Pallidum_R | 90 | Temporal_Inf_R | −3.239 |
Appendix B
| Brain Regions (Harvard–Oxford atlas) | p Values | ||
|---|---|---|---|
| Nodal Degree | Nodal Betweenness | Nodal Efficiency | |
| Young survivors of ALL > HC | |||
| 52 | 0.0434 * | 0.0212 * | 0.0252 * |
| 53 | 0.0494 * | 0.1052 | 0.0422 * |
| 71 | 0.0184 * | 0.0702 | 0.0116 * |
| 86 | 0.0254 * | 0.4185 | 0.0340 * |
| 97 | 0.0040 ** | 0.6598 | 0.0996 |
| 98 | 0.0018 ** | 0.6667 | 0.1879 |
| 99 | 0.0400 * | 0.2471 | 0.0416 * |
| 111 | 0.0222 * | 0.1581 | 0.0224 * |
| 112 | 0.0002 *** | 0.0052 ** | 0.0006 *** |
| Young survivors of ALL < HC | |||
| 3 | 0.4637 | 0.0160 * | 0.3685 |
| 4 | 0.1681 | 0.0112 * | 0.2557 |
| 11 | 0.0766 | 0.0116 * | 0.0488 * |
| 22 | 0.0254 * | 0.0084 ** | 0.0294 * |
| 39 | 0.0276 * | 0.3669 | 0.0567 |
| 43 | 0.0408 * | 0.1963 | 0.0784 |
| 49 | 0.0378 * | 0.1078 | 0.0438 * |
| 69 | 0.0766 | 0.0489 * | 0.0248 * |
| 76 | 0.2835 | 0.0299 * | 0.3667 |
| 77 | 0.3937 | 0.0449 * | 0.4637 |
| 87 | 0.0198 * | 0.0128 * | 0.0182 * |
| 96 | 0.4337 | 0.0004 *** | 0.3671 |
| 100 | 0.2157 | 0.0056 ** | 0.1478 |
| 102 | 0.3601 | 0.0392 * | 0.3973 |
| 105 | 0.0356 * | 0.0026 ** | 0.0052 ** |
| 106 | 0.1079 | 0.3445 | 0.0052 ** |
| 109 | 0.0066 ** | 0.0298 * | 0.0444 * |
| 110 | 0.0156 * | 0.1482 | 0.0804 |
References
- Neaga, A.; Jimbu, L.; Mesaros, O.; Bota, M.; Lazar, D.; Cainap, S.; Blag, C.; Zdrenghea, M. Why do children with acute lymphoblastic leukemia fare better than adults? Cancers 2021, 13, 3886. [Google Scholar] [CrossRef]
- van der Plas, E.; Modi, A.J.; Li, C.K.; Krull, K.R.; Cheung, Y.T. Cognitive impairment in survivors of pediatric acute lymphoblastic leukemia treated with chemotherapy only. J. Clin. Oncol. 2021, 39, 1705–1717. [Google Scholar] [CrossRef] [PubMed]
- Zeller, B.; Tamnes, C.K.; Kanellopoulos, A.; Amlien, I.K.; Andersson, S.; Due-Tønnessen, P.; Fjell, A.M.; Walhovd, K.B.; Westlye, L.T.; Ruud, E. Reduced neuroanatomic volumes in long-term survivors of childhood acute lymphoblastic leukemia. J. Clin. Oncol. 2013, 31, 2078–2085. [Google Scholar] [CrossRef] [PubMed]
- Tamnes, C.K.; Zeller, B.; Amlien, I.K.; Kanellopoulos, A.; Andersson, S.; Due-Tønnessen, P.; Ruud, E.; Walhovd, K.B.; Fjell, A.M. Cortical surface area and thickness in adult survivors of pediatric acute lymphoblastic leukemia. Pediatr. Blood Cancer 2015, 62, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Edelmann, M.N.; Krull, K.R.; Liu, W.; Glass, J.O.; Ji, Q.; Ogg, R.J.; Sabin, N.D.; Srivastava, D.K.; Robison, L.L.; Hudson, M.M.; et al. Diffusion tensor imaging and neurocognition in survivors of childhood acute lymphoblastic leukaemia. Brain 2014, 137, 2973–2983. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhan, Y.; He, F.; Zhang, S.; Wu, L.; Gong, H.; Zhou, F.; Zeng, X.; Xu, H. Altered functional connectivity density in young survivors of acute lymphoblastic leukemia using resting-state fMRI. Cancer Manag. Res. 2020, 12, 7033–7041. [Google Scholar] [CrossRef]
- Kesler, S.R.; Gugel, M.; Pritchard-Berman, M.; Lee, C.; Kutner, E.; Hosseini, S.M.; Dahl, G.; Lacayo, N. Altered resting state functional connectivity in young survivors of acute lymphoblastic leukemia. Pediatr. Blood Cancer 2014, 61, 1295–1299. [Google Scholar] [CrossRef]
- Kesler, S.R.; Ogg, R.; Reddick, W.E.; Phillips, N.; Scoggins, M.; Glass, J.O.; Cheung, Y.T.; Pui, C.H.; Robison, L.L.; Hudson, M.M.; et al. Brain network connectivity and executive function in long-term survivors of childhood acute lymphoblastic leukemia. Brain Connect. 2018, 8, 333–342. [Google Scholar] [CrossRef]
- Zou, L.; Su, L.; Qi, R.; Bao, F.; Fang, X.; Wang, L.; Zhai, Z.; Li, D.; Zheng, S. Abnormal topological organization in white matter structural networks in survivors of acute lymphoblastic leukaemia with chemotherapy treatment. Oncotarget 2017, 8, 60568–60575. [Google Scholar] [CrossRef]
- Wang, L.; Zou, L.; Chen, Q.; Su, L.; Xu, J.; Zhao, R.; Shan, Y.; Zhang, Q.; Zhai, Z.; Gong, X.; et al. Gray matter structural network disruptions in survivors of acute lymphoblastic leukemia with chemotherapy treatment. Acad. Radiol. 2020, 27, e27–e34. [Google Scholar] [CrossRef]
- Tijms, B.M.; Series, P.; Willshaw, D.J.; Lawrie, S.M. Similarity-based extraction of individual networks from gray matter MRI scans. Cereb. Cortex 2012, 22, 1530–1541. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jin, X.; Zhang, Y.; Wang, J. Single-subject morphological brain networks: Connectivity mapping, topological characterization and test-retest reliability. Brain Behav. 2016, 6, e00448. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.Z.; Liu, Z.; Huang, L.; Wang, X.; Yang, Z.; Zhou, G.; Zhen, Z.; Liu, J. Mapping individual brain networks using statistical similarity in regional morphology from MRI. PLoS ONE 2015, 10, e0141840. [Google Scholar]
- Yu, K.; Wang, X.; Li, Q.; Zhang, X.; Li, X.; Li, S. Individual morphological brain network construction based on multivariate euclidean distances between brain regions. Front. Hum. Neurosci. 2018, 12, 204. [Google Scholar] [CrossRef]
- Cui, L.; Li, Z.G.; Chai, Y.H.; Yu, J.; Gao, J.; Zhu, X.F.; Jin, R.M.; Shi, X.D.; Zhang, L.P.; Gao, Y.J.; et al. Outcome of children with newly diagnosed acute lymphoblastic leukemia treated with CCLG-ALL 2008: The first nation-wide prospective multicenter study in China. Am. J. Hematol. 2018, 93, 913–920. [Google Scholar] [CrossRef]
- Chen, Y.; Lei, D.; Cao, H.; Niu, R.; Chen, F.; Chen, L.; Zhou, J.; Hu, X.; Huang, X.; Guo, L.; et al. Altered single-subject gray matter structural networks in drug-naive attention deficit hyperactivity disorder children. Hum. Brain Mapp. 2021, 43, 1256–1264. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Xia, M.; Liao, X.; Evans, A.; He, Y. GRETNA: A graph theoretical network analysis toolbox for imaging connectomics. Front. Hum. Neurosci. 2015, 9, 386. [Google Scholar] [PubMed]
- Li, X.; Lei, D.; Niu, R.; Li, L.; Suo, X.; Li, W.; Yang, C.; Yang, T.; Ren, J.; Pinaya, W.H.L.; et al. Disruption of gray matter morphological networks in patients with paroxysmal kinesigenic dyskinesia. Hum. Brain Mapp. 2021, 42, 398–411. [Google Scholar] [CrossRef]
- Kong, X.Z.; Wang, X.; Huang, L.; Pu, Y.; Yang, Z.; Dang, X.; Zhen, Z.; Liu, J. Measuring individual morphological relationship of cortical regions. J. Neurosci. Methods 2014, 237, 103–107. [Google Scholar] [CrossRef]
- Genschaft, M.; Huebner, T.; Plessow, F.; Ikonomidou, V.N.; Abolmaali, N.; Krone, F.; Hoffmann, A.; Holfeld, E.; Vorwerk, P.; Kramm, C.; et al. Impact of chemotherapy for childhood leukemia on brain morphology and function. PLoS ONE 2013, 8, e78599. [Google Scholar] [CrossRef]
- Kesler, S.R.; Tanaka, H.; Koovakkattu, D. Cognitive reserve and brain volumes in pediatric acute lymphoblastic leukemia. Brain Imaging Behav. 2010, 4, 256–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, Y.T.; Krull, K.R. Neurocognitive outcomes in long-term survivors of childhood acute lymphoblastic leukemia treated on contemporary treatment protocols: A systematic review. Neurosci. Biobehav. Rev. 2015, 53, 108–120. [Google Scholar]
- Godoy, P.B.G.; Simionato, N.M.; de Mello, C.B.; Suchecki, D. Assessment of executive functions after treatment of childhood acute lymphoid leukemia: A systematic review. Neuropsychol. Rev. 2020, 30, 386–406. [Google Scholar] [CrossRef] [PubMed]
- Triarhou, L.C. Cytoarchitectonics of the rolandic operculum: Morphofunctional ponderings. Brain Struct. Func. 2021, 226, 941–950. [Google Scholar] [CrossRef]
- Hodgson, K.D.; Hutchinson, A.D.; Wilson, C.J.; Nettelbeck, T. A meta-analysis of the effects of chemotherapy on cognition in patients with cancer. Cancer Treat. Rev. 2013, 39, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Mavrea, K.; Efthymiou, V.; Katsibardi, K.; Tsarouhas, K.; Kanaka-Gantenbein, C.; Spandidos, D.A.; Chrousos, G.; Kattamis, A.; Bacopoulou, F. Cognitive function of children and adolescent survivors of acute lymphoblastic leukemia: A meta-analysis. Oncol. Lett. 2021, 21, 262. [Google Scholar] [CrossRef]
- Power, J.D.; Cohen, A.L.; Nelson, S.M.; Wig, G.S.; Barnes, K.A.; Church, J.A.; Vogel, A.C.; Laumann, T.O.; Miezin, F.M.; Schlaggar, B.L.; et al. Functional network organization of the human brain. Neuron 2011, 72, 665–678. [Google Scholar] [CrossRef] [Green Version]

| ALL Survivors | HCs | p Values | |
|---|---|---|---|
| Sex (male/female) | 16/7 | 10/10 | 0.319 a |
| Handedness (right/left) | 22/1 | 19/1 | 0.919 a |
| Age (years) | 10.3 ± 3.17 (6–16) | 9.65 ± 2.28 (7–15) | 0.587 b |
| Off-therapy duration (years) | 2.89 ± 1.43 (1–6) | n.a. | n.a. |
| Intelligence quotient (WAIS-IV) | 78.35 ± 10.4 | n.a. | n.a. |
| Trail Making Test A (s) | 94.17 ± 19.7 | n.a. | n.a. |
| Digit Span Test | 4.13 ± 1.06 | n.a. | n.a. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, Y.; Qian, L.; Wu, L.; Chen, L.; He, F.; Zhang, S.; Cheung, G.L.; Zhou, F.; Gong, H. Is Brain Network Efficiency Reduced in Young Survivors of Acute Lymphoblastic Leukemia?—Evidence from Individual-Based Morphological Brain Network Analysis. J. Clin. Med. 2022, 11, 5362. https://doi.org/10.3390/jcm11185362
Zhuang Y, Qian L, Wu L, Chen L, He F, Zhang S, Cheung GL, Zhou F, Gong H. Is Brain Network Efficiency Reduced in Young Survivors of Acute Lymphoblastic Leukemia?—Evidence from Individual-Based Morphological Brain Network Analysis. Journal of Clinical Medicine. 2022; 11(18):5362. https://doi.org/10.3390/jcm11185362
Chicago/Turabian StyleZhuang, Ying, Long Qian, Lin Wu, Linglong Chen, Fei He, Shouhua Zhang, Gerald L. Cheung, Fuqing Zhou, and Honghan Gong. 2022. "Is Brain Network Efficiency Reduced in Young Survivors of Acute Lymphoblastic Leukemia?—Evidence from Individual-Based Morphological Brain Network Analysis" Journal of Clinical Medicine 11, no. 18: 5362. https://doi.org/10.3390/jcm11185362






