Invasive and Noninvasive Nonfunctioning Gonadotroph Pituitary Tumors Differ in DNA Methylation Level of LINE-1 Repetitive Elements
Abstract
1. Introduction
2. Experimental Section
2.1. Patients and Samples
2.2. Analysis of LINE-1 DNA Methylation Level
2.3. Analysis of Genome-Wide Methylation
2.4. Reverse Transcription and Quantitative PCR
2.5. Cell Culture and Treatment
2.6. Immunohistochemical Staining
2.7. Statistical Analysis
3. Results
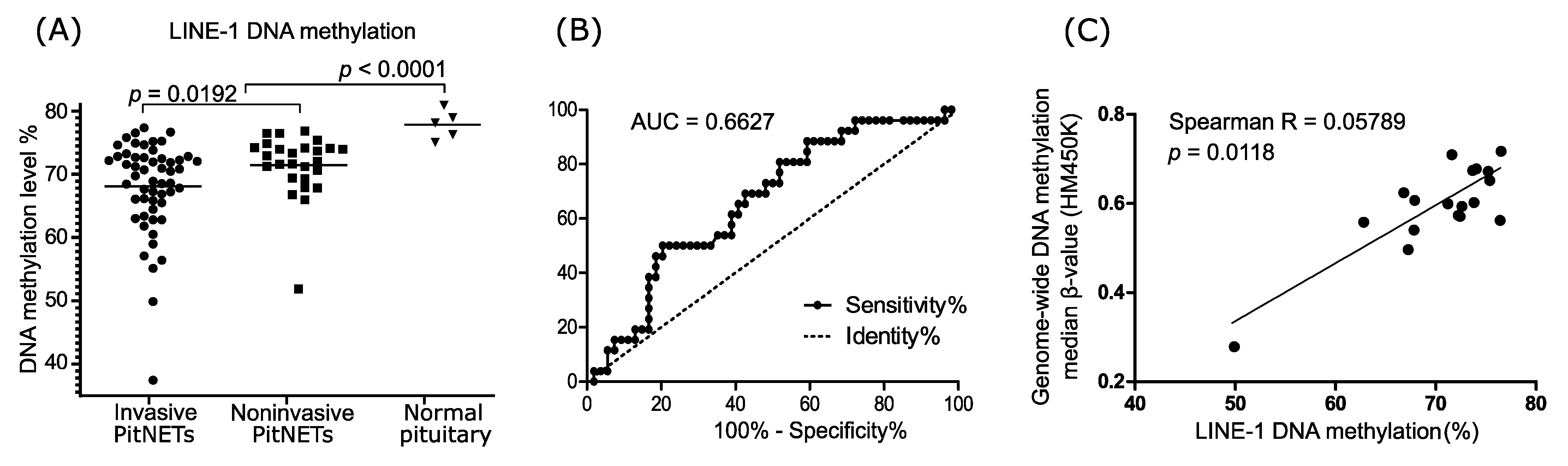
3.1. LINE-1 DNA Methylation in Nonfunctioning Gonadotroph PitNETs and Normal Pituitary
3.2. LINE-1 and Overall DNA Methylation Determined with HM450K Microarrays
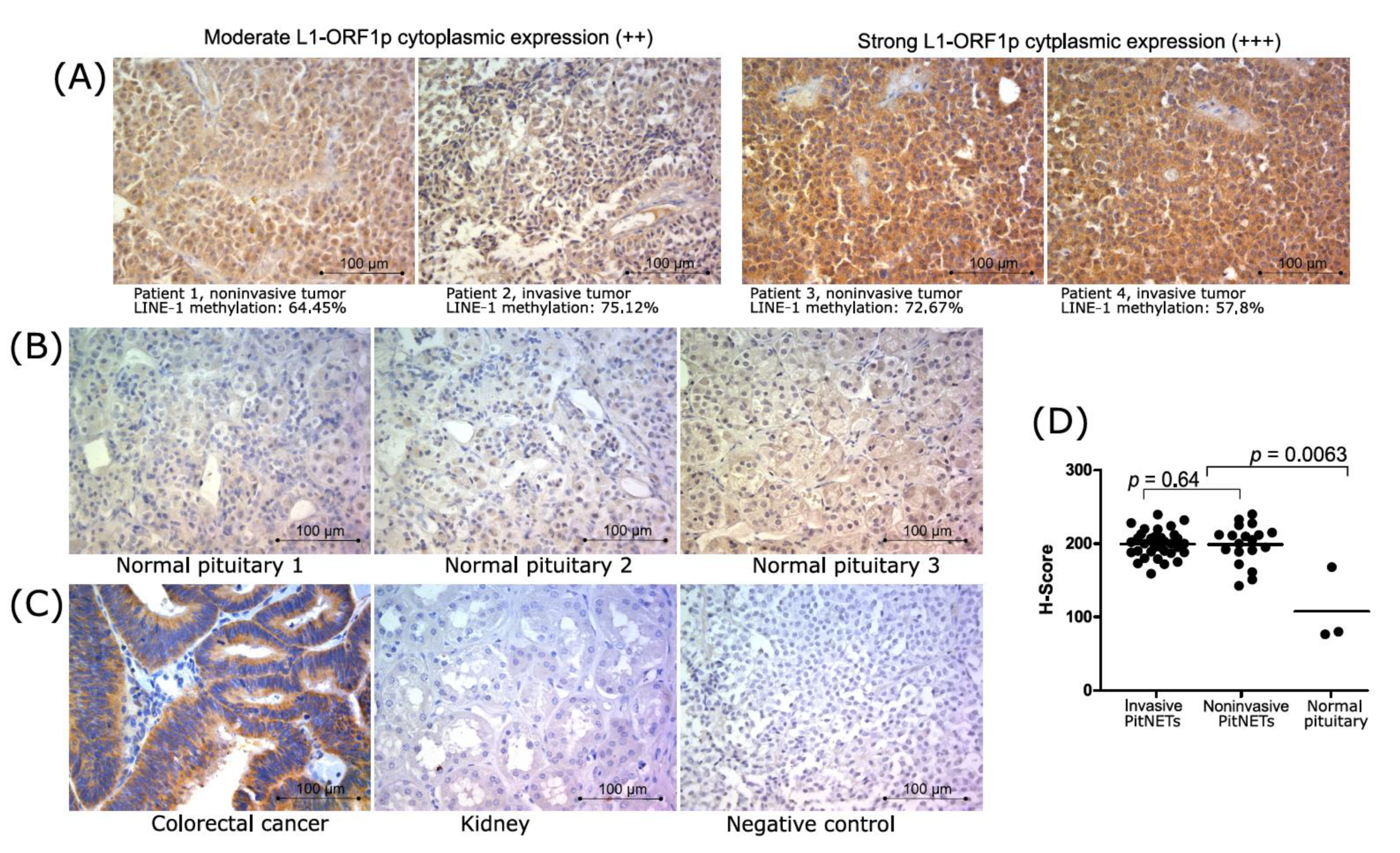
3.3. LINE-1 DNA Methylation and Expression of LINE-1 Open Reading Frame
3.4. L1-ORF1p Expression in Nonfunctioning Gonadotroph Tumor Tissue and Normal Pituitary Glands
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Greenman, Y.; Naftali, S. Non-functioning pituitary adenomas. Best Pract. Res. Clin. Endocrinol. Metab. 2009, 23, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Molitch, M.E. Diagnosis and treatment of pituitary adenomas: A review. JAMA J. Am. Med. Assoc. 2017, 317, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Mercado, M.; Melgar, V.; Salame, L.; Cuenca, D. Clinically non-functioning pituitary adenomas: Pathogenic, diagnostic and therapeutic aspects. Endocrinol. Diabetes Nutr. 2017, 64, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Raverot, G.; Vasiljevic, A.; Jouanneau, E. Prognostic factors of regrowth in nonfunctioning pituitary tumors. Pituitary 2018, 21, 176–182. [Google Scholar] [CrossRef]
- Zheng, X.; Li, S.; Zhang, W.; Zang, Z.; Hu, J.; Yang, H. Current biomarkers of invasive sporadic pituitary adenomas. Ann. Endocrinol. 2016, 77, 658–667. [Google Scholar] [CrossRef]
- Kober, P.; Boresowicz, J.; Rusetska, N.; Maksymowicz, M.; Goryca, K.; Kunicki, J.; Bonicki, W.; Siedlecki, J.A.; Bujko, M. DNA methylation profiling in nonfunctioning pituitary adenomas. Mol. Cell. Endocrinol. 2018, 473, 194–204. [Google Scholar] [CrossRef]
- Ling, C.; Pease, M.; Shi, L.; Punj, V.; Shiroishi, M.S.; Commins, D.; Weisenberger, D.J.; Wang, K.; Zada, G. A pilot genome-scale profiling of DNA methylation in sporadic pituitary macroadenomas: Association with tumor invasion and histopathological subtype. PLoS ONE 2014, 9, e96178. [Google Scholar] [CrossRef]
- Gu, Y.; Zhou, X.; Hu, F.; Yu, Y.; Xie, T.; Huang, Y.; Zhao, X.; Zhang, X. Differential DNA methylome profiling of nonfunctioning pituitary adenomas suggesting tumour invasion is correlated with cell adhesion. J. Neurooncol. 2016, 129, 23–31. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, 1–36. [Google Scholar]
- Chen, R.Z.; Pettersson, U.; Beard, C.; Jackson-Grusby, L.; Jaenisch, R. DNA hypomethylation leads to elevated mutation rates. Nature 1998, 395, 89–93. [Google Scholar] [CrossRef]
- Lander, S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [PubMed]
- Hancks, D.C.; Kazazian, H.H. Active human retrotransposons: Variation and disease. Curr. Opin. Genet. Dev. 2012, 22, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.H. Transposable elements in cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Seo, A.N.; Jung, H.Y.; Gwak, J.M.; Jung, N.; Cho, N.Y.; Kang, G.H. Alu and LINE-1 hypomethylation is associated with HER2 enriched subtype of breast cancer. PLoS ONE 2014, 9, e100429. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Nosho, K.; Kirkner, G.J.; Kawasaki, T.; Chan, A.T.; Schernhammer, E.S.; Giovannucci, E.L.; Fuchs, C.S. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J. Natl. Cancer Inst. 2008, 100, 1734–1738. [Google Scholar] [CrossRef]
- Baba, Y.; Yagi, T.; Sawayama, H.; Hiyoshi, Y.; Ishimoto, T.; Iwatsuki, M.; Miyamoto, Y.; Yoshida, N.; Baba, H. Long Interspersed Element-1 Methylation Level as a Prognostic Biomarker in Gastrointestinal Cancers. Digestion 2018, 97, 26–30. [Google Scholar] [CrossRef]
- Zelic, R.; Fiano, V.; Grasso, C.; Zugna, D.; Pettersson, A.; Gillio-Tos, A.; Merletti, F.; Richiardi, L. Global DNA hypomethylation in prostate cancer development and progression: A systematic review. Prostate Cancer Prostatic Dis. 2015, 18, 1–12. [Google Scholar] [CrossRef]
- Ohka, F.; Natsume, A.; Motomura, K.; Kishida, Y.; Kondo, Y.; Abe, T.; Nakasu, Y.; Namba, H.; Wakai, K.; Fukui, T.; et al. The Global DNA Methylation Surrogate LINE-1 Methylation Is Correlated with MGMT Promoter Methylation and Is a Better Prognostic Factor for Glioma. PLoS ONE 2011, 6, e23332. [Google Scholar] [CrossRef]
- Kawano, H.; Saeki, H.; Kitao, H.; Tsuda, Y.; Otsu, H.; Ando, K.; Ito, S.; Egashira, A.; Oki, E.; Morita, M.; et al. Chromosomal Instability Associated with Global DNA Hypomethylation is Associated with the Initiation and Progression of Esophageal Squamous Cell Carcinoma. Ann. Surg. Oncol. 2014, 21, 696–702. [Google Scholar] [CrossRef]
- Pattamadilok, J.; Huapai, N.; Rattanatanyong, P.; Vasurattana, A.; Triratanachat, S.; Tresukosol, D.; Mutirangura, A. LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int. J. Gynecol. Cancer 2008, 18, 711–717. [Google Scholar] [CrossRef]
- Tahara, T.; Tahara, S.; Horiguchi, N.; Kawamura, T.; Okubo, M.; Yamada, H.; Yoshida, D.; Ohmori, T.; Maeda, K.; Komura, N.; et al. Methylation status of IGF2 DMR and LINE1 in leukocyte DNA provides distinct clinicopathological features of gastric cancer patients. Clin. Exp. Med. 2018, 18, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Micko, A.S.G.; Wöhrer, A.; Wolfsberger, S.; Knosp, E. Invasion of the cavernous sinus space in pituitary adenomas: Endoscopic verification and its correlation with an MRI-based classification. J. Neurosurg. 2015, 122, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Knosp, E.; Steiner, E.; Kitz, K.; Matula, C.; Parent, A.D.; Laws, E.R.; Ciric, I. Pituitary adenomas with invasion of the cavernous sinus space: A magnetic resonance imaging classification compared with surgical findings. Neurosurgery 1993, 33, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Bujko, M.; Musialik, E.; Olbromski, R.; Przestrzelska, M.; Libura, M.; Pastwińska, A.; Juszczyński, P.; Zwierzchowski, L.; Baranowski, P.; Siedlecki, J.A. Repetitive genomic elements and overall DNA methylation changes in acute myeloid and childhood B-cell lymphoblastic leukemia patients. Int. J. Hematol. 2014, 100, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Bujko, M.; Rusetska, N.; Mikula, M. Validating candidate reference genes for qRT-PCR-based gene expression analysis in nonfunctioning pituitary adenomas. Pituitary 2015, 19, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Papasotiriou, I.; Pantopikou, K.; Apostolou, P. L1 retrotransposon expression in circulating tumor cells. PLoS ONE 2017, 12, e0171466. [Google Scholar] [CrossRef] [PubMed]
- Montoya-Durango, D.E.; Liu, Y.; Teneng, I.; Kalbfleisch, T.; Lacy, M.E.; Steffen, M.C.; Ramos, K.S. Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2009, 665, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Newkirk, S.J.; Lee, S.; Grandi, F.C.; Gaysinskaya, V.; Rosser, J.M.; Berg, N.V.; Hogarth, C.A.; Marchetto, M.C.N.; Muotri, A.R.; Griswold, M.D.; et al. Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc. Natl. Acad. Sci. USA 2017, 114, E5635–E5644. [Google Scholar] [CrossRef]
- Thomas, P.; Mellon, P.L.; Turgeon, J.L.; Waring, D.W. The LβT2 clonal gonadotrope: A model for single cell studies of endocrine cell secretion. Endocrinology 1996, 137, 2979–2989. [Google Scholar] [CrossRef]
- Alarid, E.T.; Windle, J.J.; Whyte, D.B.; Mellon, P.L. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development 1996, 122, 3319–3329. [Google Scholar]
- McCarty, K.S.; Szabo, E.; Flowers, J.L.; Cox, E.B.; Leight, G.S.; Miller, L.; Konrath, J.; Soper, J.T.; Budwit, D.A.; Creasman, W.T. Use of a monoclonal anti-estrogen receptor antibody in the immunohistochemical evaluation of human tumors. Cancer Res. 1986, 46, 4244–4249. [Google Scholar]
- Rodić, N.; Sharma, R.; Sharma, R.; Zampella, J.; Dai, L.; Taylor, M.S.; Hruban, R.H.; Iacobuzio-Donahue, C.A.; Maitra, A.; Torbenson, M.S.; et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 2014, 184, 1280–1286. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.M.; Shin, S.H.; Kwon, H.J.; Park, S.Y.; Kook, M.C.; Kim, Y.W.; Cho, N.Y.; Kim, N.; Kim, T.Y.; Kim, D.; et al. ALU and LINE-1 hypomethylations in multistep gastric carcinogenesis and their prognostic implications. Int. J. Cancer 2012, 131, 1323–1331. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, F.; Hodgson, J.G.; Eden, A.; Jackson-Grusby, L.; Dausman, J.; Gray, J.W.; Leonhardt, H.; Jaenisch, R. Induction of tumors in mice by genomic hypomethylation. Science 2003, 300, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Yeung, C.M.; Chan, C.B.; Leung, P.S.; Cheng, C.H.K. Cells of the anterior pituitary. Int. J. Biochem. Cell Biol. 2006, 38, 1441–1449. [Google Scholar]
- Neou, M.; Villa, C.; Armignacco, R.; Jouinot, A.; Raffin-Sanson, M.L.; Septier, A.; Letourneur, F.; Diry, S.; Diedisheim, M.; Izac, B.; et al. Pangenomic Classification of Pituitary Neuroendocrine Tumors. Cancer Cell 2020, 37, 123–134.e5. [Google Scholar] [CrossRef]
- Wolff, E.M.; Byun, H.M.; Han, H.F.; Sharma, S.; Nichols, P.W.; Siegmund, K.D.; Yang, A.S.; Jones, P.A.; Liang, G. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010, 6, e1000917. [Google Scholar] [CrossRef]
- Ardeljan, D.; Taylor, M.S.; Ting, D.T.; Burns, K.H. The human long interspersed element-1 retrotransposon: An emerging biomarker of Neoplasia. Clin. Chem. 2017, 63, 816–822. [Google Scholar] [CrossRef]
- Laverrière, J.N.; L’Hôte, D.; Tabouy, L.; Schang, A.L.; Quérat, B.; Cohen-Tannoudji, J. Epigenetic regulation of alternative promoters and enhancers in progenitor, immature, and mature gonadotrope cell lines. Mol. Cell. Endocrinol. 2016, 434, 250–265. [Google Scholar] [CrossRef]
- Xie, H.; Hoffmann, H.M.; Iyer, A.K.; Brayman, M.J.; Ngo, C.; Sunshine, M.J.; Mellon, P.L. Chromatin status and transcription factor binding to gonadotropin promoters in gonadotrope cell lines. Reprod. Biol. Endocrinol. 2017, 15, 86. [Google Scholar] [CrossRef]
- Yosefzon, Y.; David, C.; Tsukerman, A.; Pnueli, L.; Qiao, S.; Boehm, U.; Melamed, P. An epigenetic switch repressing Tet1 in gonadotropes activates the reproductive axis. Proc. Natl. Acad. Sci. USA 2017, 114, 201704393. [Google Scholar] [CrossRef]
- Shademan, M.; Zare, K.; Zahedi, M.; Mosannen Mozaffari, H.; Bagheri Hosseini, H.; Ghaffarzadegan, K.; Goshayeshi, L.; Dehghani, H. Promoter methylation, transcription, and retrotransposition of LINE-1 in colorectal adenomas and adenocarcinomas. Cancer Cell Int. 2020, 20, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Emery, S.B.; Flasch, D.A.; Wang, Y.; Kwan, K.Y.; Kidd, J.M.; Moran, J.V.; Mills, R.E. Identification and characterization of occult human-specific LINE-1 insertions using long-read sequencing technology. Nucleic Acids Res. 2020, 48, 1146–1163. [Google Scholar] [CrossRef]
- Harris, C.R.; Normart, R.; Yang, Q.; Stevenson, E.; Haffty, B.G.; Ganesan, S.; Cordon-Cardo, C.; Levine, A.J.; Tang, L.H. Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Genes Cancer 2010, 1, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Sur, D.; Kustwar, R.K.; Budania, S.; Mahadevan, A.; Hancks, D.C.; Yadav, V.; Shankar, S.K.; Mandal, P.K. Detection of the LINE-1 retrotransposon RNA-binding protein ORF1p in different anatomical regions of the human brain. Mob. DNA 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Horn, A.V.; Han, J.S. Transposons and Retrotransposons. In Methods in Molecular Biology; Springer Science+Business Media: New York, NY, USA, 2016; Volume 1400, pp. 131–137. ISBN 978-1-4939-3370-9. [Google Scholar]
- Rodriguez-Martin, B.; Alvarez, E.G.; Baez-Ortega, A.; Zamora, J.; Supek, F.; Demeulemeester, J.; Santamarina, M.; Ju, Y.S.; Temes, J.; Garcia-Souto, D.; et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat. Genet. 2020, 52, 306–319. [Google Scholar] [CrossRef]
- Kinzler, K.W.; Vogelstein, B.; Horii, A.; Miyoshi, Y.; Nakamura, Y. Disruption of the APC Gene by a Retrotransposal Insertion of LI Sequence in a Colon Cancer. Cancer Res. 1992, 52, 643–645. [Google Scholar]
- Aschacher, T.; Wolf, B.; Enzmann, F.; Kienzl, P.; Messner, B.; Sampl, S.; Svoboda, M.; Mechtcheriakova, D.; Holzmann, K.; Bergmann, M. LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines. Oncogene 2016, 35, 94–104. [Google Scholar] [CrossRef] [PubMed]

| Nonfunctioning Gonadotroph PitNET Patients (Number of Patients) | 80 |
|---|---|
| Demographical data | |
| Age (years) | |
| Median; range | 59.5; 34–82 |
| Gender | |
| Male | 46 |
| Female | 34 |
| Histopathology | |
| Gonadotroph tumors | 79 |
| Plurihormonal (TSH, FSH, LH, α-subunit) | 1 |
| Alphoma type (α-subunit positive only) | 3 |
| Null cell tumor/ultrastructurally gonadotroph | 7 |
| Tumors with oncocytic features | 17 |
| Ki67 index > 3% | 5 |
| Clinical classification | |
| Invasive PitNET | 54 |
| Noninvasive PitNET | 26 |
| Tumor size > 10 mm | 66 |
| Tumor size < 10 mm | 14 |
| Newly diagnosed | 68 |
| Recurrent | 12 |
| Recurrence during follow-up | 10 |
| Weak Expression | Moderate Expression | High Expression | |
|---|---|---|---|
| Normal pituitary | 2 | 1 | 0 |
| Invasive tumors (n = 17) | 0 | 16 | 18 |
| Noninvasive tumors (n = 13) | 0 | 7 | 12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rusetska, N.; Kober, P.; Król, S.K.; Boresowicz, J.; Maksymowicz, M.; Kunicki, J.; Bonicki, W.; Bujko, M. Invasive and Noninvasive Nonfunctioning Gonadotroph Pituitary Tumors Differ in DNA Methylation Level of LINE-1 Repetitive Elements. J. Clin. Med. 2021, 10, 560. https://doi.org/10.3390/jcm10040560
Rusetska N, Kober P, Król SK, Boresowicz J, Maksymowicz M, Kunicki J, Bonicki W, Bujko M. Invasive and Noninvasive Nonfunctioning Gonadotroph Pituitary Tumors Differ in DNA Methylation Level of LINE-1 Repetitive Elements. Journal of Clinical Medicine. 2021; 10(4):560. https://doi.org/10.3390/jcm10040560
Chicago/Turabian StyleRusetska, Natalia, Paulina Kober, Sylwia Katarzyna Król, Joanna Boresowicz, Maria Maksymowicz, Jacek Kunicki, Wiesław Bonicki, and Mateusz Bujko. 2021. "Invasive and Noninvasive Nonfunctioning Gonadotroph Pituitary Tumors Differ in DNA Methylation Level of LINE-1 Repetitive Elements" Journal of Clinical Medicine 10, no. 4: 560. https://doi.org/10.3390/jcm10040560
APA StyleRusetska, N., Kober, P., Król, S. K., Boresowicz, J., Maksymowicz, M., Kunicki, J., Bonicki, W., & Bujko, M. (2021). Invasive and Noninvasive Nonfunctioning Gonadotroph Pituitary Tumors Differ in DNA Methylation Level of LINE-1 Repetitive Elements. Journal of Clinical Medicine, 10(4), 560. https://doi.org/10.3390/jcm10040560






