Heavy Atom Detergent/Lipid Combined X-ray Crystallography for Elucidating the Structure-Function Relationships of Membrane Proteins
Abstract
:1. Introduction
2. Heavy Atom Labeled Protein Ligands
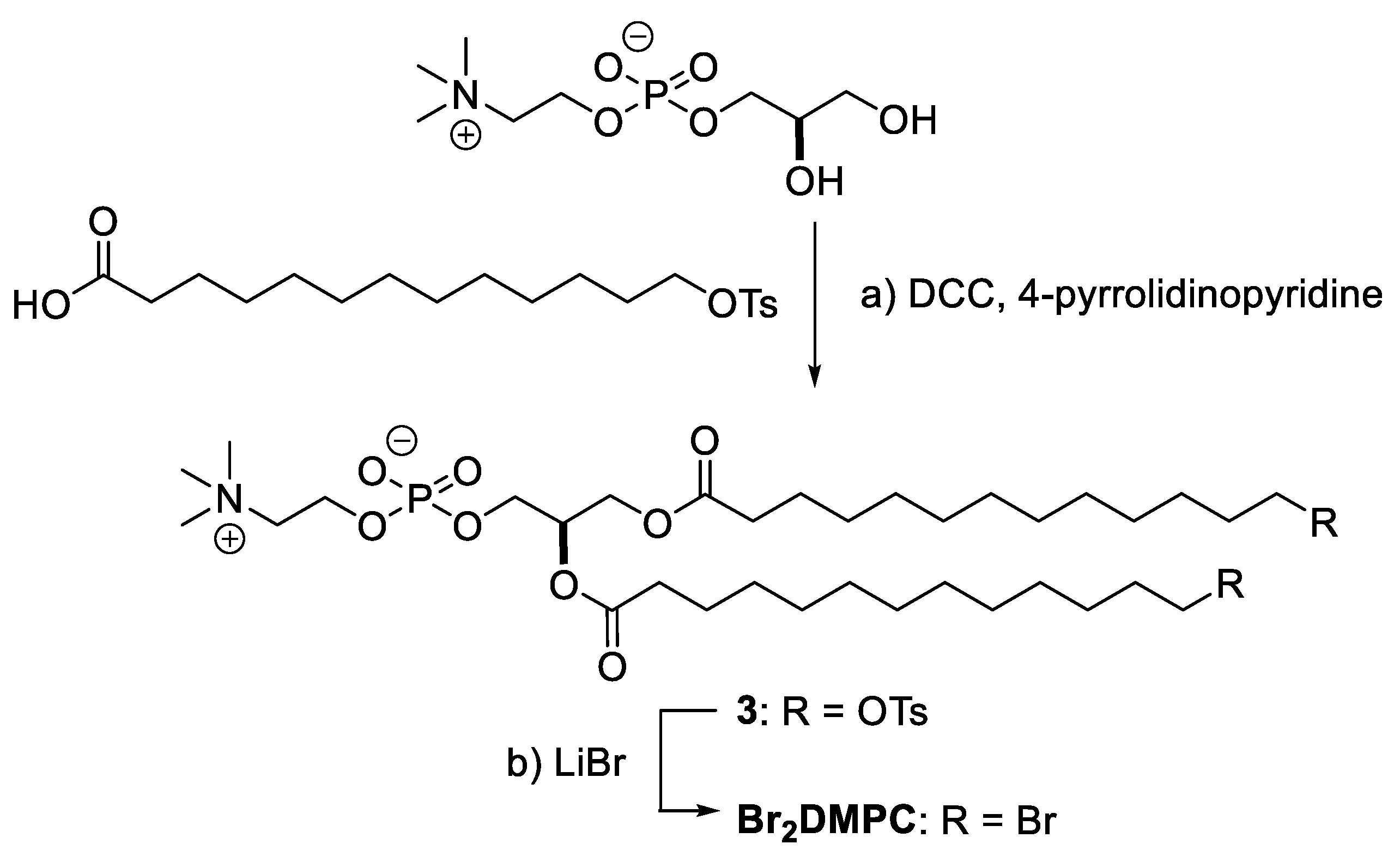
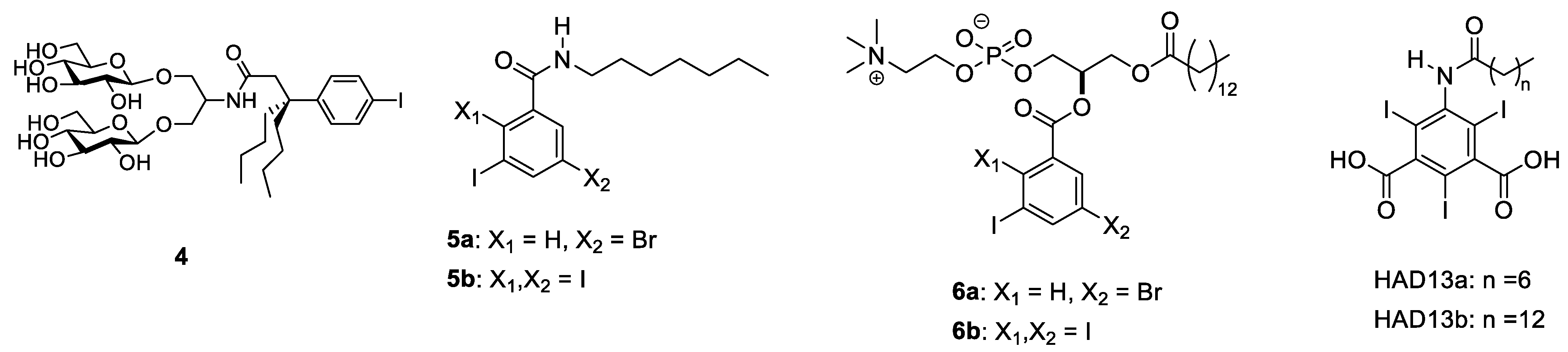
2.1. Halogens
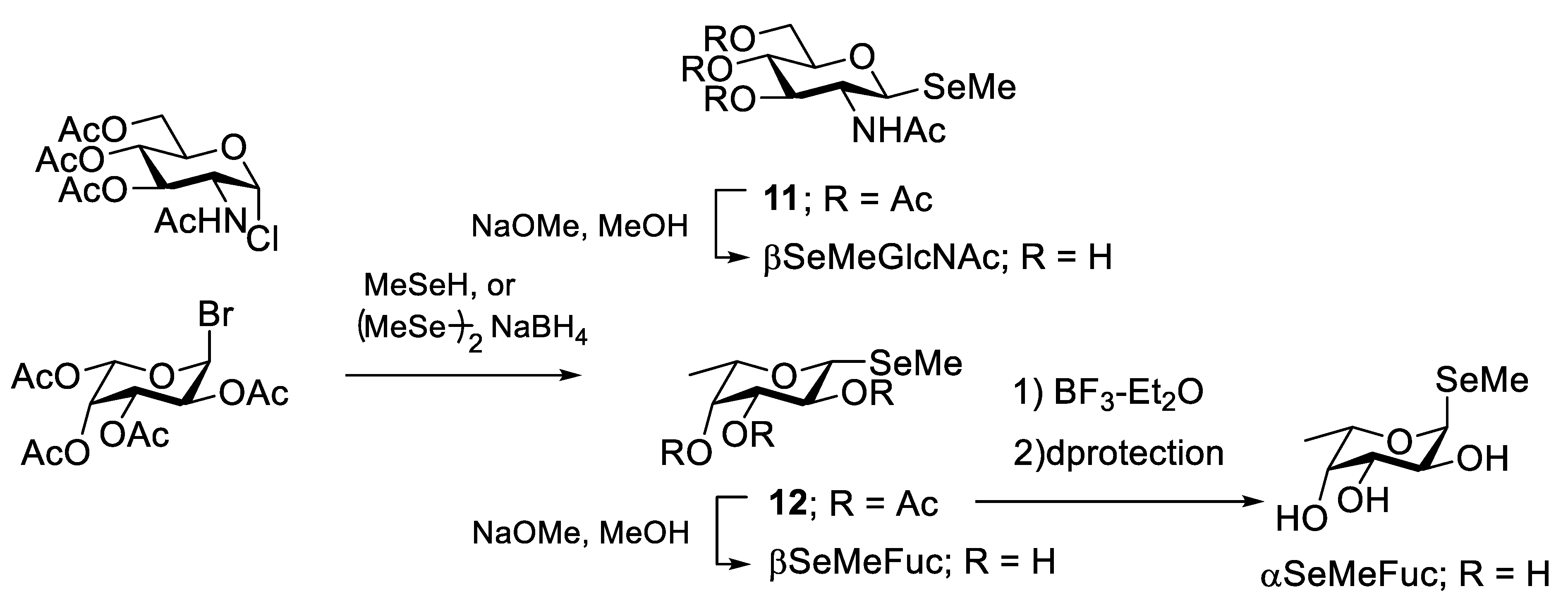
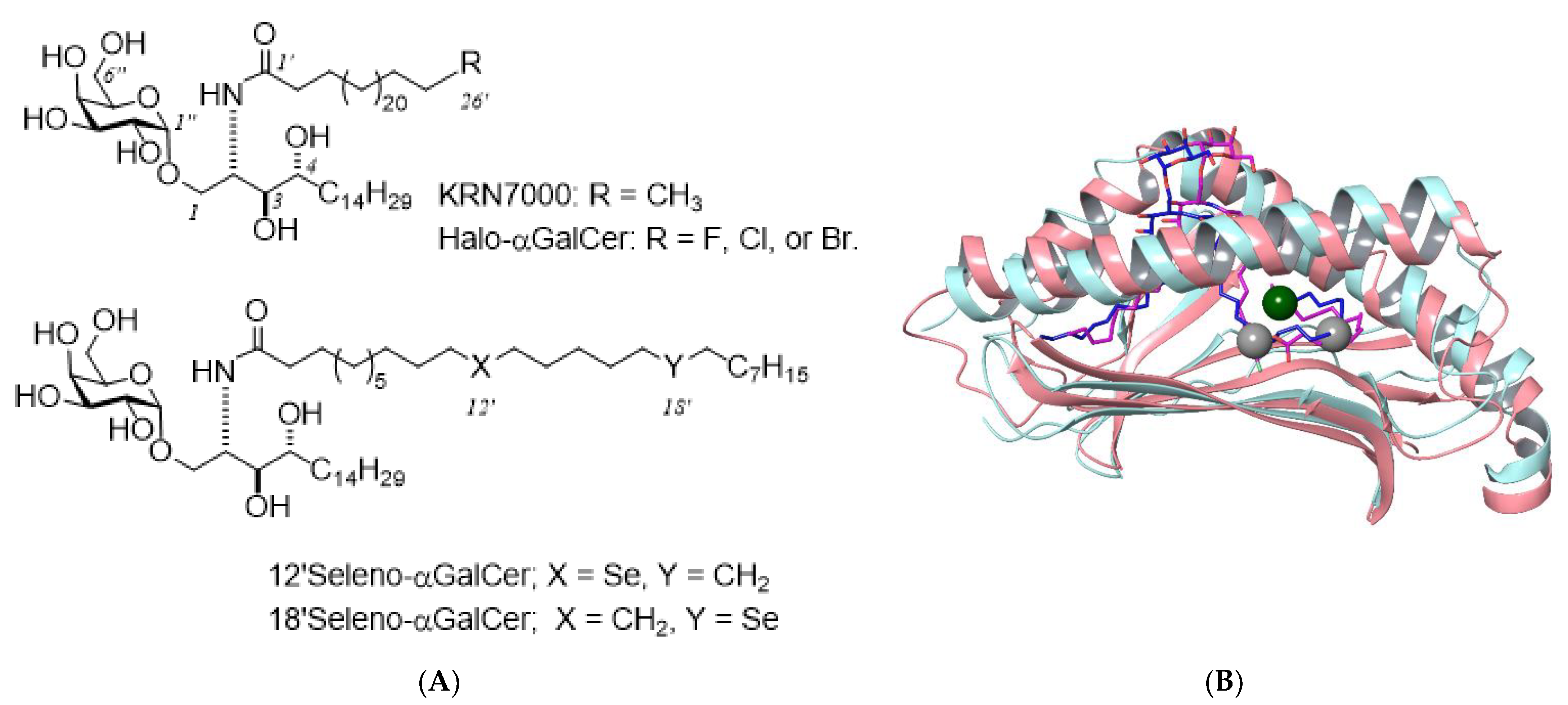
2.2. Selenium
3. Serial Femtosecond Crystallography (SFX)
3.1. De Novo Phasing in SFX
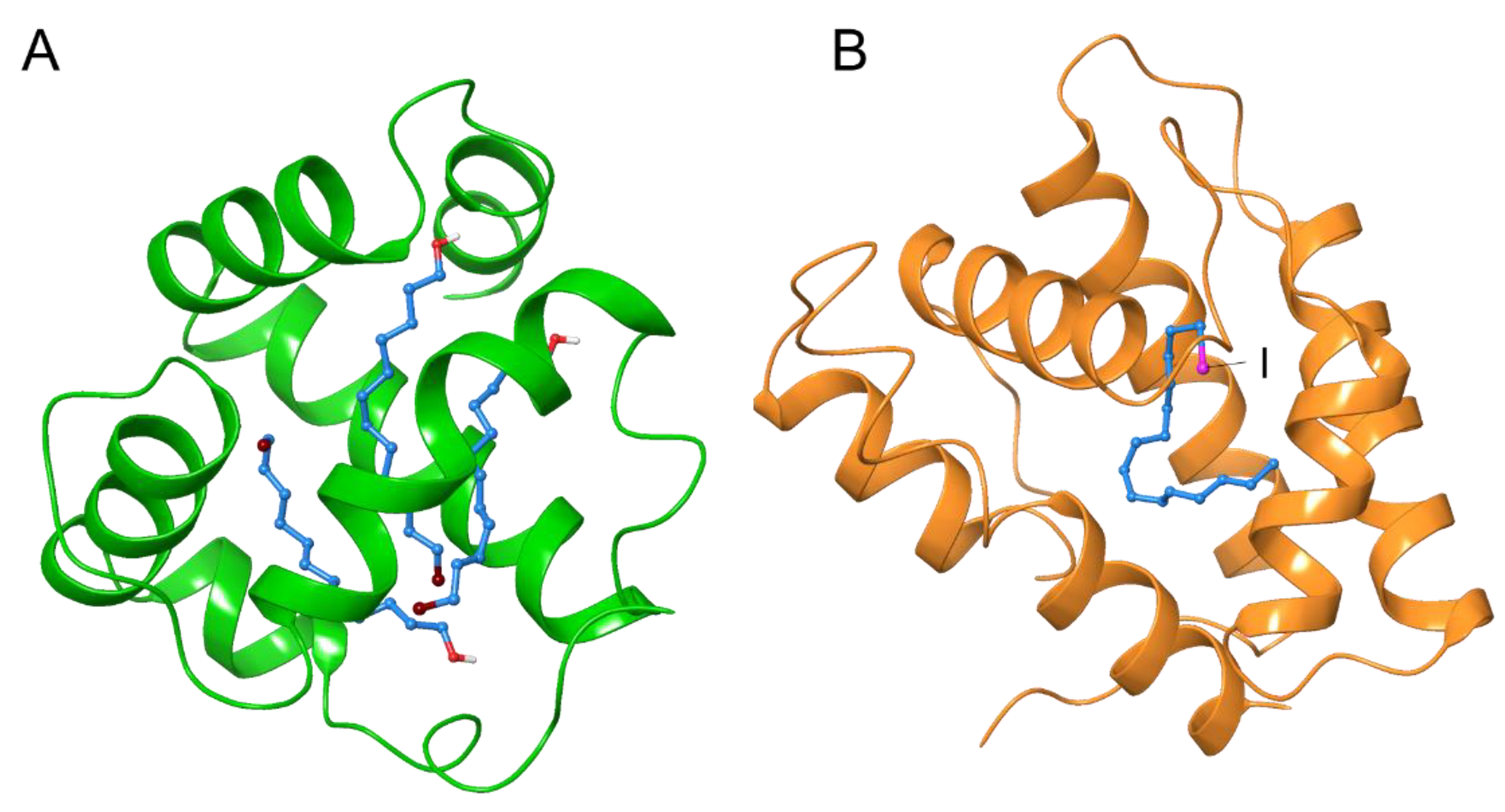
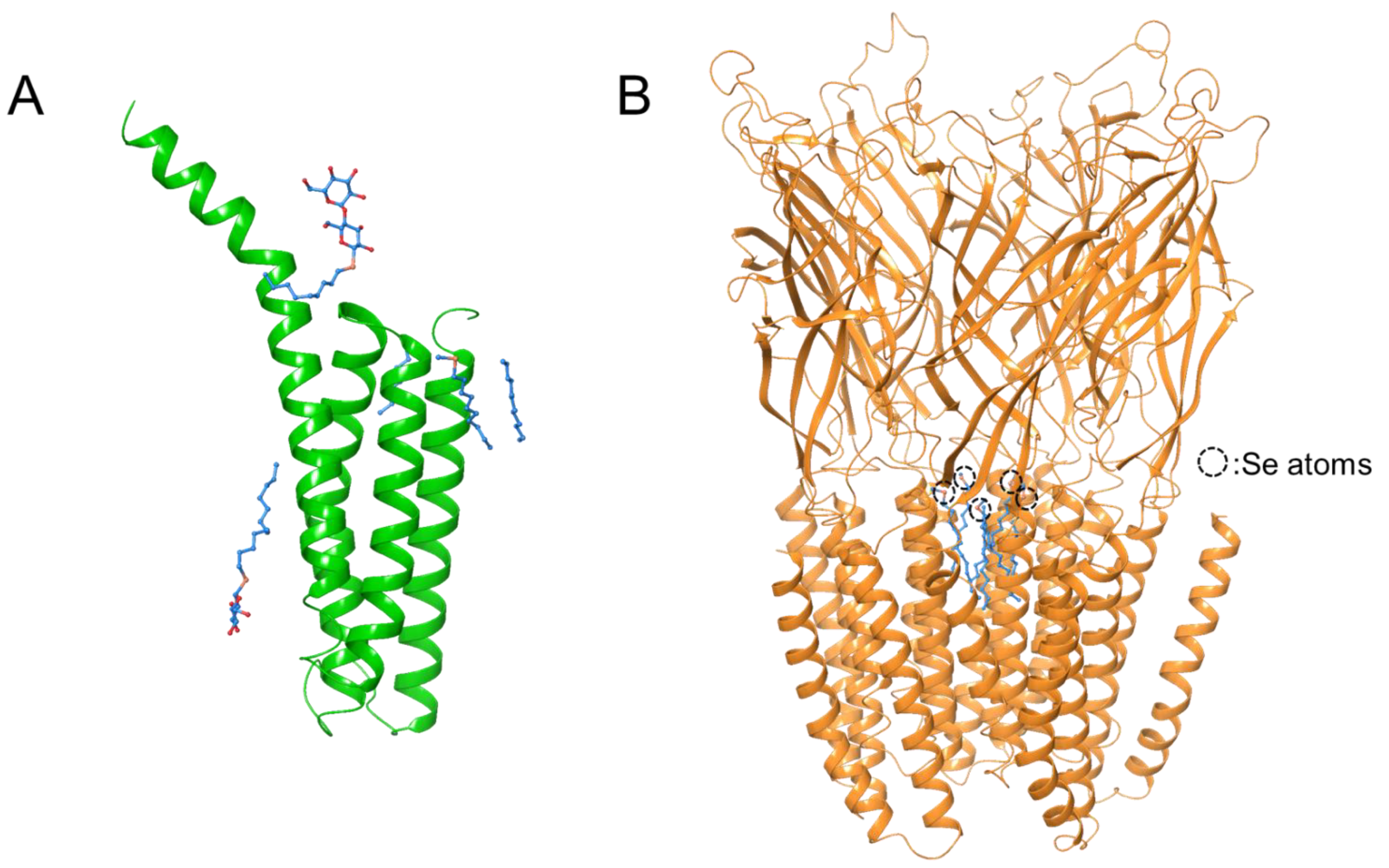
3.2. De Novo Phasing with the HAD13a Detergent
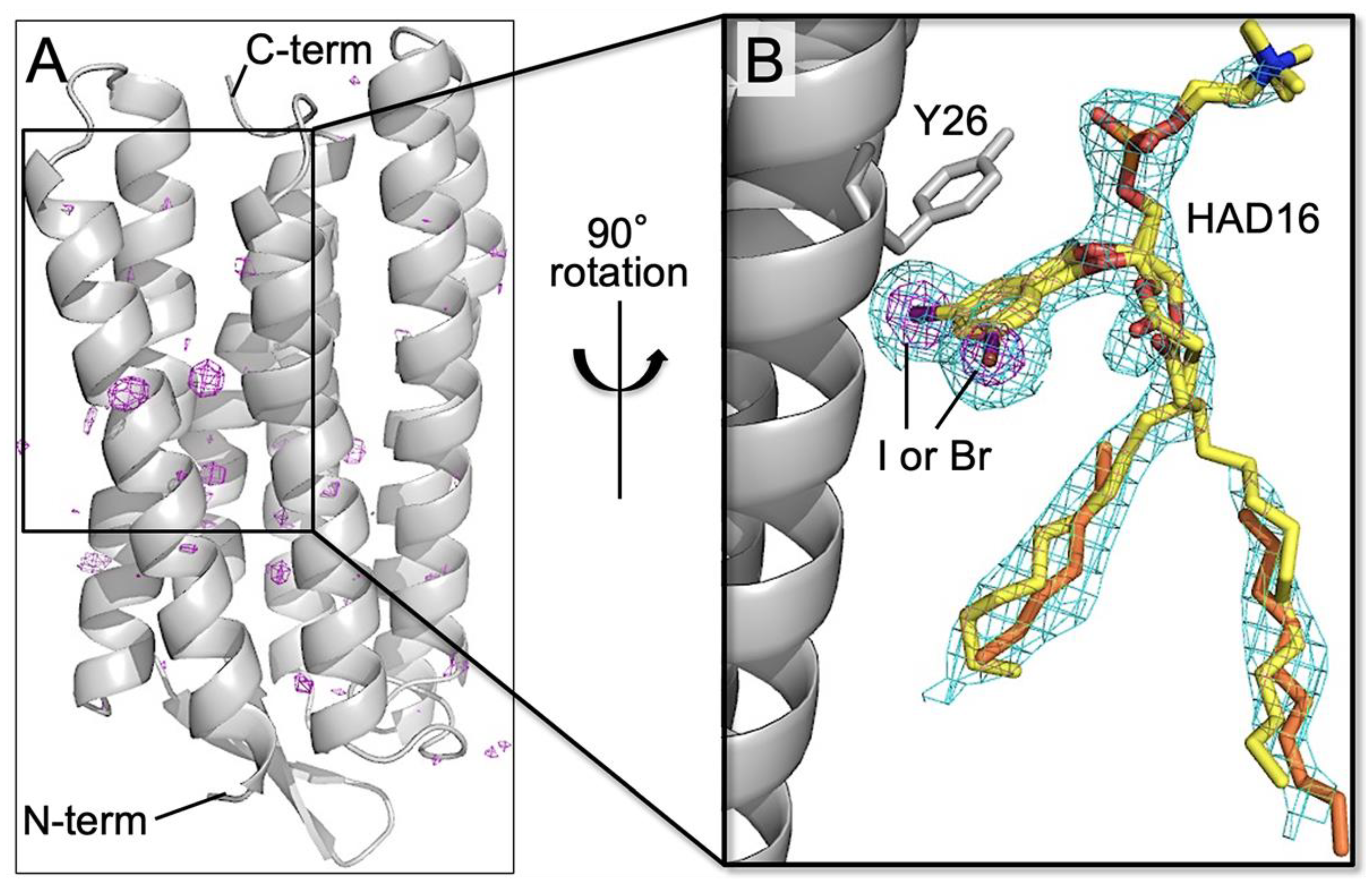
3.3. Binding of the HAD16 Lipid to a Membrane Protein
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A2A GPCR | G protein-coupled A2a adenosine receptor |
| α-GalCer | α-galactosylceramide |
| β-MeSe-Fuc | selenomethyl derivative of fucose |
| βMeSe-GlcNAc | Selenomethyl-N-acetylglucosamine |
| CHAPSO | 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxypropanesulfonate |
| CMC | critical micelle concentration |
| DDM | dodecyl-β-D-maltoside |
| DMPC | 1,2-dimyristoyl-sn-glycero-3-phosphocholine |
| DPPC | dipalmitoylphosphatidylcholine |
| ERATO | Exploratory Research for Advanced Technology |
| I3C | 5-amino-2,4,6-triiodoisophthalic acid |
| JST | Japan Science and Technology Agency |
| LCLS | Linac Coherent Light Source |
| LCP | lipidic cubic phase |
| MAD | multi-wavelength anomalous diffraction |
| MEXT | Ministry of Education, Culture, Sports, Science and Technology |
| MIR | multiple heavy-atom isomorphous replacement |
| MIRAS | multiple heavy-atom isomorphous replacement with anomalous scattering |
| PAL-XFEL | Pohang Accelerator Laboratory X-ray Free Electron Laser |
| PDB | Protein Data Bank |
| PRESTO | Precursory Research for Embryonic Science and Technology |
| SACLA | SPring-8 Angstrom Compact Free Electron Laser |
| SAD | single-wavelength anomalous diffraction |
| SeDDM | dodecyl-β-D-selenomaltoside |
| SFX | serial femtosecond crystallography |
| SIR | single heavy-atom isomorphous replacement |
| SIRAS | single heavy-atom isomorphous replacement with anomalous scattering |
| SRX | synchrotron radiation crystallography |
| SwissFEL | Swiss X-ray Free Electron Laser |
| XFEL | X-ray free-electron laser |
References
- Murata, M.; Sugiyama, S.; Matsuoka, S.; Matsumori, N. Bioactive Structure of Membrane Lipids and Natural Products Elucidated by a Chemistry-Based Approach. Chem. Rec. 2015, 15, 675–690. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Mizohata, E.; Nagatoishi, S.; Watanabe, T.; Nakakido, M.; Iwanari, H.; Mochizuki, Y.; Nakayama, T.; Kado, Y.; Yokota, Y.; et al. Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface. Structure 2019, 27, 519–527.e515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayama, T.; Mizohata, E.; Yamashita, T.; Nagatoishi, S.; Nakakido, M.; Iwanari, H.; Mochizuki, Y.; Kado, Y.; Yokota, Y.; Satoh, R.; et al. Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses. Protein Sci. A Publ. Protein Soc. 2015, 24, 328–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugiyama, A.; Kawamura, T.; Tanaka, T.; Doi, H.; Yamashita, T.; Shinoda, K.; Fujitani, H.; Yamatsugu, K.; Shimizu, Y.; Tatsumi, T.; et al. Cupid and Psyche system for the diagnosis and treatment of advanced cancer. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2019, 95, 602–611. [Google Scholar] [CrossRef] [Green Version]
- Kado, Y.; Mizohata, E.; Nagatoishi, S.; Iijima, M.; Shinoda, K.; Miyafusa, T.; Nakayama, T.; Yoshizumi, T.; Sugiyama, A.; Kawamura, T.; et al. Epiregulin recognition mechanisms by anti-epiregulin antibody 9E5: Structural, functional, and molecular dynamics simulation analyses. J. Biol. Chem. 2016, 291, 2319–2330. [Google Scholar] [CrossRef] [Green Version]
- Uchihashi, T.; Ganser, C. Recent advances in bioimaging with high-speed atomic force microscopy. Biophys. Rev. 2020, 12, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Renard, K.; Byrne, B. Insights into the Role of Membrane Lipids in the Structure, Function and Regulation of Integral Membrane Proteins. Int. J. Mol. Sci. 2021, 22, 9026. [Google Scholar] [CrossRef] [PubMed]
- Mizohata, E.; Nakane, T.; Fukuda, Y.; Nango, E.; Iwata, S. Serial femtosecond crystallography at the SACLA: Breakthrough to dynamic structural biology. Biophys. Rev. 2018, 10, 209–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendrickson, W.A.; Teeter, M.M. Structure of the Hydrophobic Protein Crambin Determined Directly from the Anomalous Scattering of Sulfur. Nature 1981, 290, 107–113. [Google Scholar] [CrossRef] [Green Version]
- Agniswamy, J.; Joyce, M.G.; Hammer, C.H.; Sun, P.D. Towards a rational approach for heavy-atom derivative screening in protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008, 64, 354–367. [Google Scholar] [CrossRef]
- Lu, J.; Sun, P.D. A rapid and rational approach to generating isomorphous heavy-atom phasing derivatives. FEBS J. 2014, 281, 4021–4028. [Google Scholar] [CrossRef] [Green Version]
- Allen, F.H.K.O.; Watson, D.G.; Brammer, L.; Orpen, A.G.; Taylor, R. Tables of bond lengths determined by x-ray and neutron diffraction. Part 1. Bond lengths in organic compounds. J. Chem. Soc. Perkin Trans. 1987, 2, 1–19. [Google Scholar] [CrossRef]
- Blanksby, S.J.; Ellison, G.B. Bond dissociation energies of organic molecules. Acc. Chem. Res. 2003, 36, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.Y.; Pike, A.C.; Mackenzie, A.; McClenaghan, C.; Aryal, P.; Dong, L.; Quigley, A.; Grieben, M.; Goubin, S.; Mukhopadhyay, S.; et al. K2P channel gating mechanisms revealed by structures of TREK-2 and a complex with Prozac. Science 2015, 347, 1256–1259. [Google Scholar] [CrossRef]
- Ulens, C.; Spurny, R.; Thompson, A.J.; Alqazzaz, M.; Debaveye, S.; Han, L.; Price, K.; Villalgordo, J.M.; Tresadern, G.; Lynch, J.W.; et al. The prokaryote ligand-gated ion channel ELIC captured in a pore blocker-bound conformation by the Alzheimer’s disease drug memantine. Structure 2014, 22, 1399–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martick, M.; Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 2006, 126, 309–320. [Google Scholar] [CrossRef] [Green Version]
- Stagno, J.R.; Yu, P.; Dyba, M.A.; Wang, Y.X.; Liu, Y. Heavy-atom labeling of RNA by PLOR for de novo crystallographic phasing. PLoS ONE 2019, 14, e0215555. [Google Scholar] [CrossRef]
- Czapinska, H.; Winiewska-Szajewska, M.; Szymaniec-Rutkowska, A.; Piasecka, A.; Bochtler, M.; Poznanski, J. Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2. J. Phys. Chem. B 2021, 125, 2491–2503. [Google Scholar] [CrossRef]
- Scholfield, M.R.; Zanden, C.M.; Carter, M.; Ho, P.S. Halogen bonding (X-bonding): A biological perspective. Protein Sci. A Publ. Protein Soc. 2013, 22, 139–152. [Google Scholar] [CrossRef]
- Silvius, J.R. Cholesterol modulation of lipid intermixing in phospholipid and glycosphingolipid mixtures. Evaluation using fluorescent lipid probes and brominated lipid quenchers. Biochemistry 1992, 31, 3398–3408. [Google Scholar] [CrossRef]
- Abrams, F.S.; London, E. Calibration of the parallax fluorescence quenching method for determination of membrane penetration depth: Refinement and comparison of quenching by spin-labeled and brominated lipids. Biochemistry 1992, 31, 5312–5322. [Google Scholar] [CrossRef] [PubMed]
- Bolen, E.J.; Holloway, P.W. Quenching of tryptophan fluorescence by brominated phospholipid. Biochemistry 1990, 29, 9638–9643. [Google Scholar] [CrossRef]
- Halter, M.; Nogata, Y.; Dannenberger, O.; Sasaki, T.; Vogel, V. Engineered lipids that cross-link the inner and outer leaflets of lipid bilayers. Langmuir 2004, 20, 2416–2423. [Google Scholar] [CrossRef] [PubMed]
- Markello, T.; Zlotnick, A.; Everett, J.; Tennyson, J.; Holloway, P.W. Determination of the topography of cytochrome b5 in lipid vesicles by fluorescence quenching. Biochemistry 1985, 24, 2895–2901. [Google Scholar] [CrossRef]
- Cudmore, A.J.; Bradshaw, J.P.; Alecio, M.R. X-Ray-Diffraction Studies Using a Novel Synthetic Phospholipid. Biophys. Chem. 1994, 49, 71–76. [Google Scholar] [CrossRef]
- Roszak, A.W.; Gardiner, A.T.; Isaacs, N.W.; Cogdell, R.J. Brominated lipids identify lipid binding sites on the surface of the reaction center from Rhodobacter sphaeroides. Biochemistry 2007, 46, 2909–2916. [Google Scholar] [CrossRef] [PubMed]
- Campanacci, V.; Lartigue, A.; Hallberg, B.M.; Jones, T.A.; Giudici-Orticoni, M.T.; Tegoni, M.; Cambillau, C. Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding. Proc. Natl. Acad. Sci. USA 2003, 100, 5069–5074. [Google Scholar] [CrossRef] [Green Version]
- Lautenschlager, C.; Leal, W.S.; Clardy, J. Bombyx mori pheromone-binding protein binding nonpheromone ligands: Implications for pheromone recognition. Structure 2007, 15, 1148–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chae, P.S.C.; Cho, K.H.; Bae, H.E. Heavy atom-bearing tripod amphiphiles for the membrane protein study. New J. Chem. 2014, 38, 2354–2361. [Google Scholar] [CrossRef]
- Beck, T.; Krasauskas, A.; Gruene, T.; Sheldrick, G.M. A magic triangle for experimental phasing of macromolecules. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008, 64, 1179–1182. [Google Scholar] [CrossRef]
- Sippel, K.H.; Robbins, A.H.; Reutzel, R.; Domsic, J.; Boehlein, S.K.; Govindasamy, L.; Agbandje-McKenna, M.; Rosser, C.J.; McKenna, R. Structure determination of the cancer-associated Mycoplasma hyorhinis protein Mh-p37. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008, 64, 1172–1178. [Google Scholar] [CrossRef] [PubMed]
- Nakane, T.; Hanashima, S.; Suzuki, M.; Saiki, H.; Hayashi, T.; Kakinouchi, K.; Sugiyama, S.; Kawatake, S.; Matsuoka, S.; Matsumori, N.; et al. Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent. Proc. Natl. Acad. Sci. USA 2016, 113, 13039–13044. [Google Scholar] [CrossRef] [Green Version]
- Katzberg, R.W. Urography into the 21st century: New contrast media, renal handling, imaging characteristics, and nephrotoxicity. Radiology 1997, 204, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.B.; Watson, A.D. Metal-Based X-ray Contrast Media. Chem. Rev. 1999, 99, 2353–2378. [Google Scholar] [CrossRef]
- Beck, T.; Gruene, T.; Sheldrick, G.M. The magic triangle goes MAD: Experimental phasing with a bromine derivative. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 374–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truong, J.Q.; Nguyen, S.; Bruning, J.B.; Shearwin, K.E. Simplified heavy-atom derivatization of protein structures via co-crystallization with the MAD tetragon tetrabromoterephthalic acid. Acta Cryst. F Struct. Biol. Commun. 2021, 77, 156–162. [Google Scholar] [CrossRef]
- Faham, S.; Boulting, G.L.; Massey, E.A.; Yohannan, S.; Yang, D.; Bowie, J.U. Crystallization of bacteriorhodopsin from bicelle formulations at room temperature. Protein Sci. A Publ. Protein Soc. 2005, 14, 836–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendrickson, W.A.; Horton, J.R.; LeMaster, D.M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): A vehicle for direct determination of three-dimensional structure. EMBO J. 1990, 9, 1665–1672. [Google Scholar] [CrossRef]
- Lin, L.; Sheng, J.; Huang, Z. Nucleic acid X-ray crystallography via direct selenium derivatization. Chem. Soc. Rev. 2011, 40, 4591–4602. [Google Scholar] [CrossRef] [PubMed]
- Merino-Montiel, P.L.; López, Ó.; Fernández-Bolaños, J.G. L-Isofucoselenofagomine and derivatives: Dual activities as antioxidants and as glycosidase inhibitors. Tetrahedron 2012, 68, 3591–3595. [Google Scholar] [CrossRef]
- Mangiavacchi, F.; Coelho Dias, I.F.; Di Lorenzo, I.; Grzes, P.; Palomba, M.; Rosati, O.; Bagnoli, L.; Marini, F.; Santi, C.; Lenardao, E.J.; et al. Sweet Selenium: Synthesis and Properties of Selenium-Containing Sugars and Derivatives. Pharmaceuticals 2020, 13. [Google Scholar] [CrossRef]
- Davies, M.J.S.; Schiesser, C.H. 1,4-Anhydro-4-seleno-d-talitol (SeTal): A remarkable selenium-containing therapeutic molecule. New J. Chem. 2019, 43, 9759–9765. [Google Scholar] [CrossRef]
- Buts, L.; Loris, R.; De Genst, E.; Oscarson, S.; Lahmann, M.; Messens, J.; Brosens, E.; Wyns, L.; De Greve, H.; Bouckaert, J. Solving the phase problem for carbohydrate-binding proteins using selenium derivatives of their ligands: A case study involving the bacterial F17-G adhesin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003, 59, 1012–1015. [Google Scholar] [CrossRef]
- Kostlanova, N.; Mitchell, E.P.; Lortat-Jacob, H.; Oscarson, S.; Lahmann, M.; Gilboa-Garber, N.; Chambat, G.; Wimmerova, M.; Imberty, A. The fucose-binding lectin from Ralstonia solanacearum. A new type of beta-propeller architecture formed by oligomerization and interacting with fucoside, fucosyllactose, and plant xyloglucan. J. Biol. Chem. 2005, 280, 27839–27849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houser, J.; Komarek, J.; Kostlanova, N.; Cioci, G.; Varrot, A.; Kerr, S.C.; Lahmann, M.; Balloy, V.; Fahy, J.V.; Chignard, M.; et al. A soluble fucose-specific lectin from Aspergillus fumigatus conidia--structure, specificity and possible role in fungal pathogenicity. PLoS ONE 2013, 8, e83077. [Google Scholar] [CrossRef] [PubMed]
- Sulak, O.; Cioci, G.; Delia, M.; Lahmann, M.; Varrot, A.; Imberty, A.; Wimmerova, M. A TNF-like trimeric lectin domain from Burkholderia cenocepacia with specificity for fucosylated human histo-blood group antigens. Structure 2010, 18, 59–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommer, R.; Hauck, D.; Varrot, A.; Imberty, A.; Kunzler, M.; Titz, A. O-Alkylated heavy atom carbohydrate probes for protein X-ray crystallography: Studies towards the synthesis of methyl 2-O-methyl-L-selenofucopyranoside. Beilstein. J. Org. Chem. 2016, 12, 2828–2833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimabukuro, J.; Makyio, H.; Suzuki, T.; Nishikawa, Y.; Kawasaki, M.; Imamura, A.; Ishida, H.; Ando, H.; Kato, R.; Kiso, M. Synthesis of seleno-fucose compounds and their application to the X-ray structural determination of carbohydrate-lectin complexes using single/multi-wavelength anomalous dispersion phasing. Bioorg. Med. Chem. 2017, 25, 1132–1142. [Google Scholar] [CrossRef] [PubMed]
- Makyio, H.; Shimabukuro, J.; Suzuki, T.; Imamura, A.; Ishida, H.; Kiso, M.; Ando, H.; Kato, R. Six independent fucose-binding sites in the crystal structure of Aspergillus oryzae lectin. Biochem. Biophys. Res. Commun. 2016, 477, 477–482. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Hayashi, C.; Komura, N.; Tamai, R.; Uzawa, J.; Ogawa, J.; Tanaka, H.N.; Imamura, A.; Ishida, H.; Kiso, M.; et al. Synthesis and Glycan-Protein Interaction Studies of Se-Sialosides by (77)Se NMR. Org. Lett. 2019, 21, 6393–6396. [Google Scholar] [CrossRef] [PubMed]
- Saino, H.; Ago, H.; Ukita, Y.; Miyano, M. Seleno-detergent MAD phasing of leukotriene C4 synthase in complex with dodecyl-beta-D-selenomaltoside. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 1666–1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauguet, L.; Poitevin, F.; Murail, S.; Van Renterghem, C.; Moraga-Cid, G.; Malherbe, L.; Thompson, A.W.; Koehl, P.; Corringer, P.J.; Baaden, M.; et al. Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. EMBO J. 2013, 32, 728–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fredga, A.L.; Lindgren, A. Selena-hexanoic and Selena-pentanoic Acids. Acta Chem. Scand. 1961, 15, 938–939. [Google Scholar] [CrossRef] [Green Version]
- Golmohammadi, R. Studies on the methods of preparation of di- and tri-selena-straight-chain fatty acids and corresponding amides, with some remarks on the infrared spectra of the mono-, di- and tri-selena-fatty acids. Acta Chem. Scand. 1966, 20, 563–571. [Google Scholar] [CrossRef]
- Sadek, S.A.B.; Basmadjian, G.P. Selenium labeled fatty acids as potential myocardial imaging agents. J. Label. Compd. Radiopharm. 1983, 20, 487–494. [Google Scholar] [CrossRef]
- Yanishlieva, N.R.; Marinova, E.; Houte, H.; Partali, V.; Sliwka, H.R. 11-Selenadodecylglyceryl-1-ether in lipid autoxidation. J. Am. Oil Chem. Soc. 2001, 78, 691–696. [Google Scholar] [CrossRef]
- Houte, H.; Partali, V.; Sliwka, H.R.; Quartey, E.G. Synthesis of structured lipids and etherlipids with antioxidants: Combination of a selena fatty acid and a selena fatty alcohol with a carotenoic acid in glyceride molecules. Chem. Phys. Lipids 2000, 105, 105–113. [Google Scholar] [CrossRef]
- Kabara, J.J.; Vrable, R. Antimicrobial lipids: Natural and synthetic fatty acids and monoglycerides. Lipids 1977, 12, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.I.; Hanashima, S.; Nomura, T.; Lethu, S.; Tsuchikawa, H.; Murata, M.; Kusaka, H.; Kita, S.; Maenaka, K. Synthesis and Th1-immunostimulatory activity of alpha-galactosylceramide analogues bearing a halogen-containing or selenium-containing acyl chain. Bioorg. Med. Chem. 2016, 24, 3687–3695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emma, P.; Akre, R.; Arthur, J.; Bionta, R.; Bostedt, C.; Bozek, J.; Brachmann, A.; Bucksbaum, P.; Coffee, R.; Decker, F.J.; et al. First lasing and operation of an angstrom-wavelength free-electron laser. Nat. Photonics 2010, 4, 641–647. [Google Scholar] [CrossRef]
- Ishikawa, T.; Aoyagi, H.; Asaka, T.; Asano, Y.; Azumi, N.; Bizen, T.; Ego, H.; Fukami, K.; Fukui, T.; Furukawa, Y.; et al. A compact X-ray free-electron laser emitting in the sub-angstrom region. Nat. Photonics 2012, 6, 540–544. [Google Scholar] [CrossRef]
- Decking, W.; Abeghyan, S.; Abramian, P.; Abramsky, A.; Aguirre, A.; Albrecht, C.; Alou, P.; Altarelli, M.; Altmann, P.; Amyan, K.; et al. A MHz-repetition-rate hard X-ray free-electron laser driven by a superconducting linear accelerator. Nat. Photonics 2020, 14, 391–397. [Google Scholar] [CrossRef]
- Kang, H.S.; Min, C.K.; Heo, H.; Kim, C.; Yang, H.; Kim, G.; Nam, I.; Baek, S.Y.; Choi, H.J.; Mun, G.; et al. Hard X-ray free-electron laser with femtosecond-scale timing jitter. Nat. Photonics 2017, 11, 708–713. [Google Scholar] [CrossRef]
- Prat, E.; Abela, R.; Aiba, M.; Alarcon, A.; Alex, J.; Arbelo, Y.; Arrell, C.; Arsov, V.; Bacellar, C.; Beard, C.; et al. A compact and cost-effective hard X-ray free-electron laser driven by a high-brightness and low-energy electron beam. Nat. Photonics 2020, 14, 748–754. [Google Scholar] [CrossRef]
- Chapman, H.N.; Fromme, P.; Barty, A.; White, T.A.; Kirian, R.A.; Aquila, A.; Hunter, M.S.; Schulz, J.; DePonte, D.P.; Weierstall, U.; et al. Femtosecond X-ray protein nanocrystallography. Nature 2011, 470, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Tono, K.; Nango, E.; Sugahara, M.; Song, C.; Park, J.; Tanaka, T.; Tanaka, R.; Joti, Y.; Kameshima, T.; Ono, S.; et al. Diverse application platform for hard X-ray diffraction in SACLA (DAPHNIS): Application to serial protein crystallography using an X-ray free-electron laser. J. Synchrotron Radiat. 2015, 22, 532–537. [Google Scholar] [CrossRef]
- Fukuda, Y.; Tse, K.M.; Nakane, T.; Nakatsu, T.; Suzuki, M.; Sugahara, M.; Inoue, S.; Masuda, T.; Yumoto, F.; Matsugaki, N.; et al. Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography. Proc. Natl. Acad. Sci. USA 2016, 113, 2928–2933. [Google Scholar] [CrossRef] [Green Version]
- Barty, A.; Caleman, C.; Aquila, A.; Timneanu, N.; Lomb, L.; White, T.A.; Andreasson, J.; Arnlund, D.; Bajt, S.; Barends, T.R.; et al. Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements. Nat. Photonics 2012, 6, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Barends, T.R.; Foucar, L.; Botha, S.; Doak, R.B.; Shoeman, R.L.; Nass, K.; Koglin, J.E.; Williams, G.J.; Boutet, S.; Messerschmidt, M.; et al. De novo protein crystal structure determination from X-ray free-electron laser data. Nature 2014, 505, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, K.; Pan, D.; Okuda, T.; Sugahara, M.; Kodan, A.; Yamaguchi, T.; Murai, T.; Gomi, K.; Kajiyama, N.; Mizohata, E.; et al. An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography. Sci. Rep. 2015, 5, 14017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakane, T.; Song, C.; Suzuki, M.; Nango, E.; Kobayashi, J.; Masuda, T.; Inoue, S.; Mizohata, E.; Nakatsu, T.; Tanaka, T.; et al. Native sulfur/chlorine SAD phasing for serial femtosecond crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 2519–2525. [Google Scholar] [CrossRef] [PubMed]
- Colletier, J.P.; Sawaya, M.R.; Gingery, M.; Rodriguez, J.A.; Cascio, D.; Brewster, A.S.; Michels-Clark, T.; Hice, R.H.; Coquelle, N.; Boutet, S.; et al. De novo phasing with X-ray laser reveals mosquito larvicide BinAB structure. Nature 2016, 539, 43–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, M.S.; Yoon, C.H.; DeMirci, H.; Sierra, R.G.; Dao, E.H.; Ahmadi, R.; Aksit, F.; Aquila, A.L.; Ciftci, H.; Guillet, S.; et al. Selenium single-wavelength anomalous diffraction de novo phasing using an X-ray-free electron laser. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Batyuk, A.; Galli, L.; Ishchenko, A.; Han, G.W.; Gati, C.; Popov, P.A.; Lee, M.Y.; Stauch, B.; White, T.A.; Barty, A.; et al. Native phasing of x-ray free-electron laser data for a G protein-coupled receptor. Sci. Adv. 2016, 2, e1600292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cogdell, R.J.; Gardiner, A.T.; Roszak, A.W.; Stoncius, S.; Kocovsky, P.; Isaacs, N.W. Mapping lipid and detergent molecules at the surface of membrane proteins. Biochem. Soc. Trans. 2011, 39, 775–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nango, E.; Royant, A.; Kubo, M.; Nakane, T.; Wickstrand, C.; Kimura, T.; Tanaka, T.; Tono, K.; Song, C.; Tanaka, R.; et al. A three-dimensional movie of structural changes in bacteriorhodopsin. Science 2016, 354, 1552–1557. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanashima, S.; Nakane, T.; Mizohata, E. Heavy Atom Detergent/Lipid Combined X-ray Crystallography for Elucidating the Structure-Function Relationships of Membrane Proteins. Membranes 2021, 11, 823. https://doi.org/10.3390/membranes11110823
Hanashima S, Nakane T, Mizohata E. Heavy Atom Detergent/Lipid Combined X-ray Crystallography for Elucidating the Structure-Function Relationships of Membrane Proteins. Membranes. 2021; 11(11):823. https://doi.org/10.3390/membranes11110823
Chicago/Turabian StyleHanashima, Shinya, Takanori Nakane, and Eiichi Mizohata. 2021. "Heavy Atom Detergent/Lipid Combined X-ray Crystallography for Elucidating the Structure-Function Relationships of Membrane Proteins" Membranes 11, no. 11: 823. https://doi.org/10.3390/membranes11110823





