Characterization of H2O2-Induced Alterations in Global Transcription of mRNA and lncRNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Viability Analysis
2.3. Hydrogen Peroxide Treatment and RNA Extraction
2.4. Sample Qualification and Quantification
2.5. Library Preparation and Sequencing
2.6. Identification of lncRNA
2.7. Quantification and Differential Expression Analysis
2.8. lncRNA Target Gene Prediction
2.9. Transcription Factor Prediction
2.10. Quantitative Reverse-Transcription Polymerase Chain Reaction
2.11. Deposit of Transcriptome Data in Public Database
3. Results
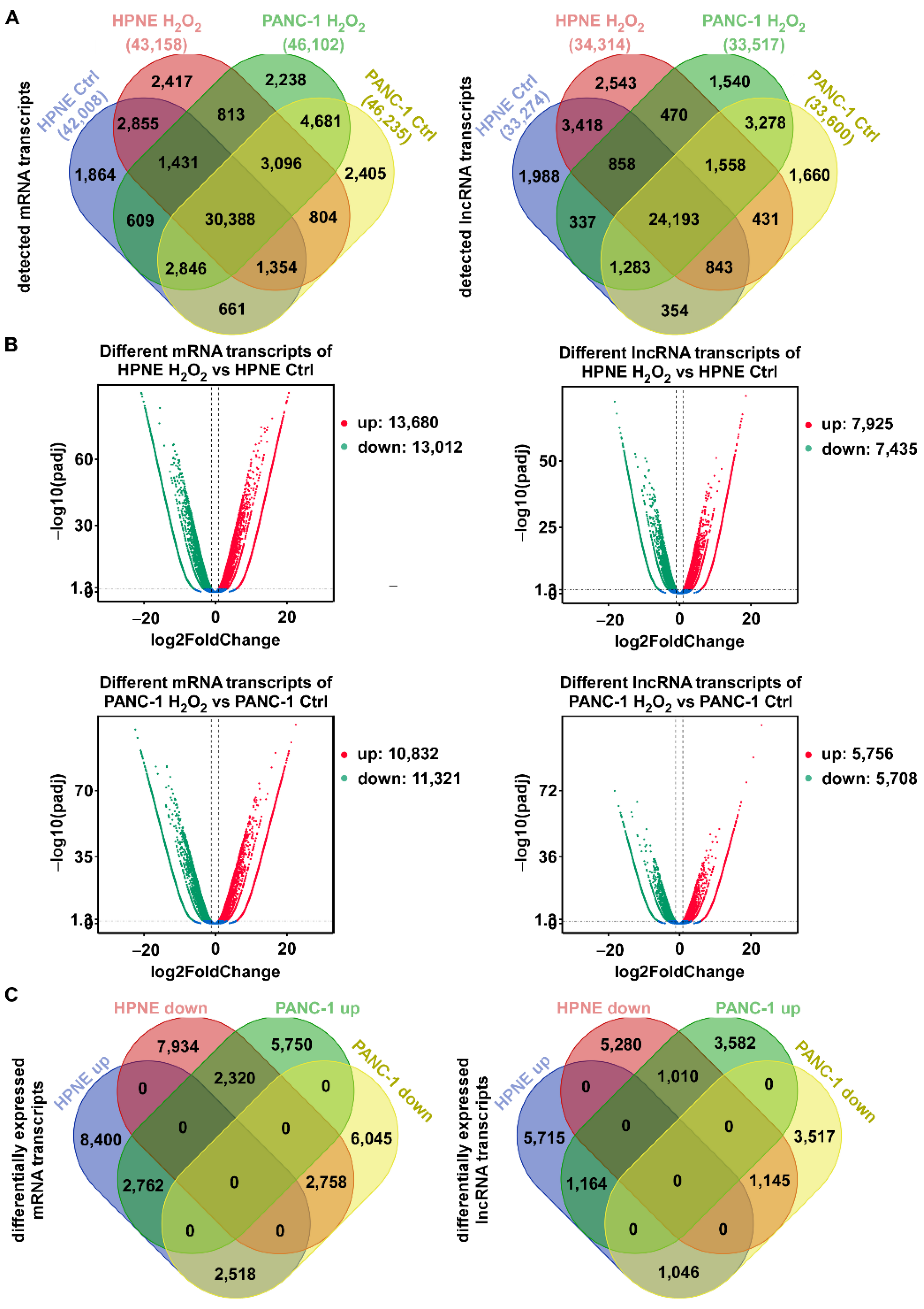
3.1. Global Changes in mRNA and lncRNA Transcripts in Response to H2O2 Stress in Normal Pancreatic Epithelial and Pancreatic Cancer Cells
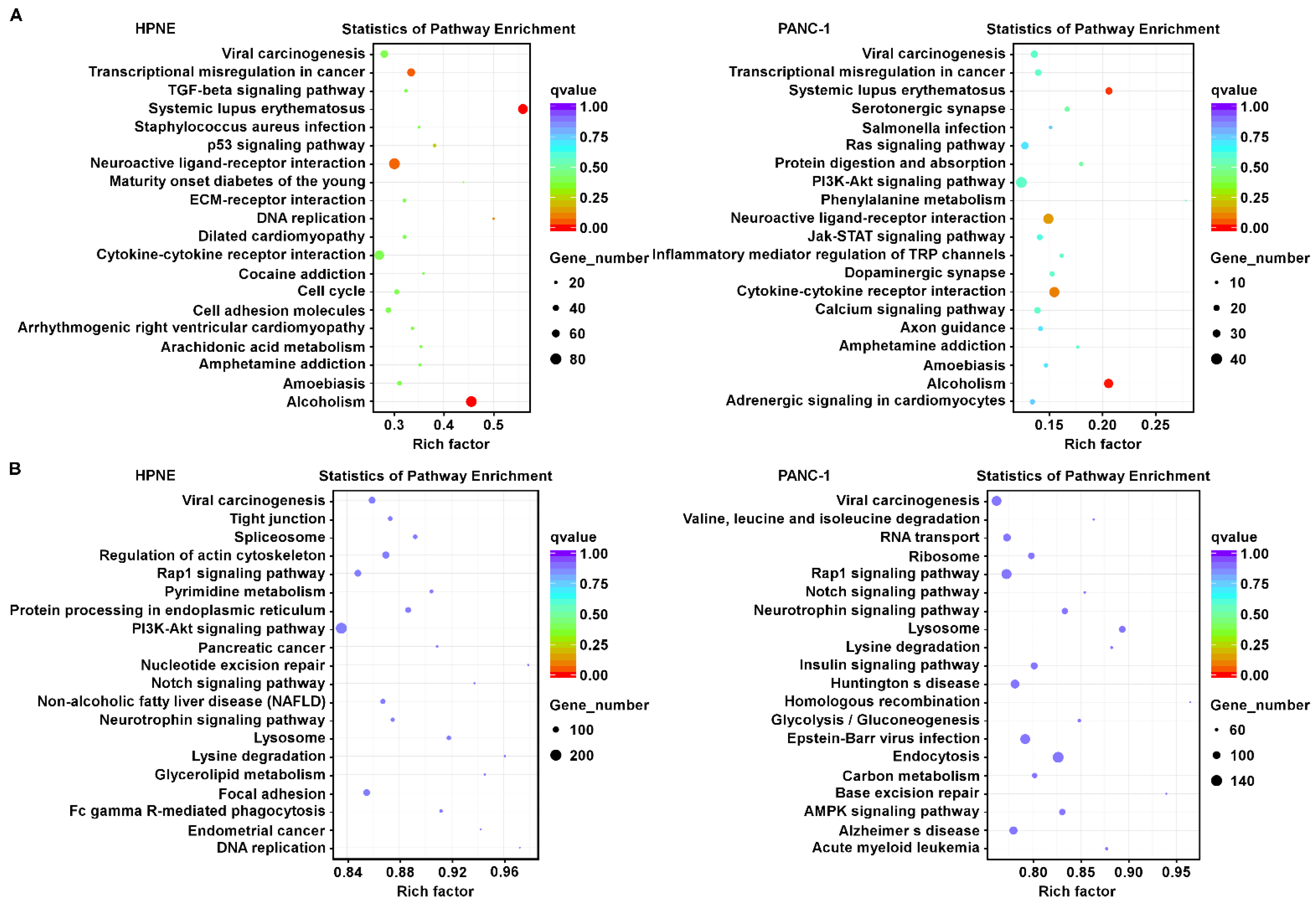
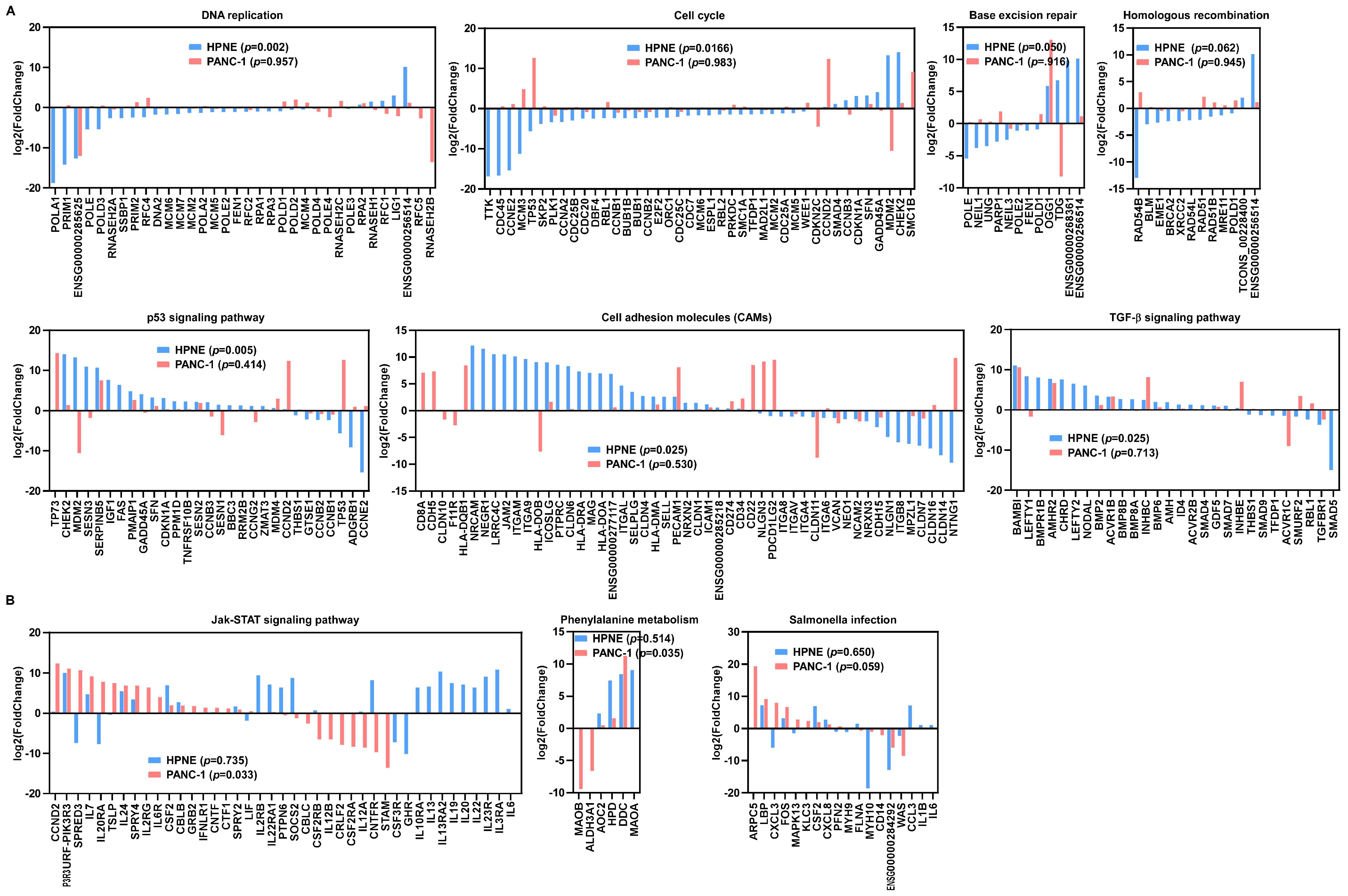
3.2. Comparison of Changes in Signal Transduction Pathways Induced by H2O2 in Pancreatic Epithelial Cells and Pancreatic Cancer Cells
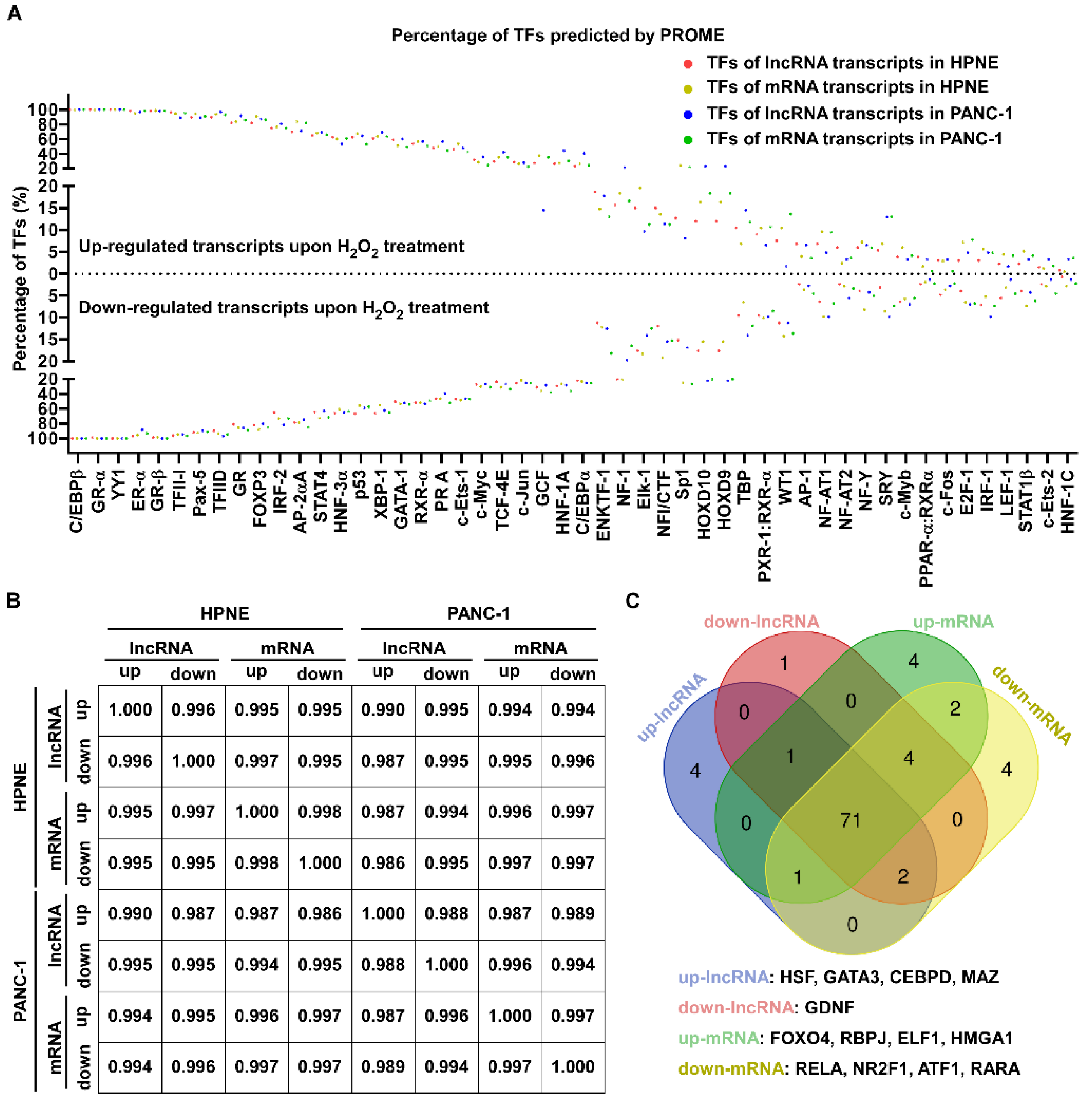
3.3. Alterations of mRNA and lncRNA Transcripts Induced by H2O2 Largely Shared the Same Promoters
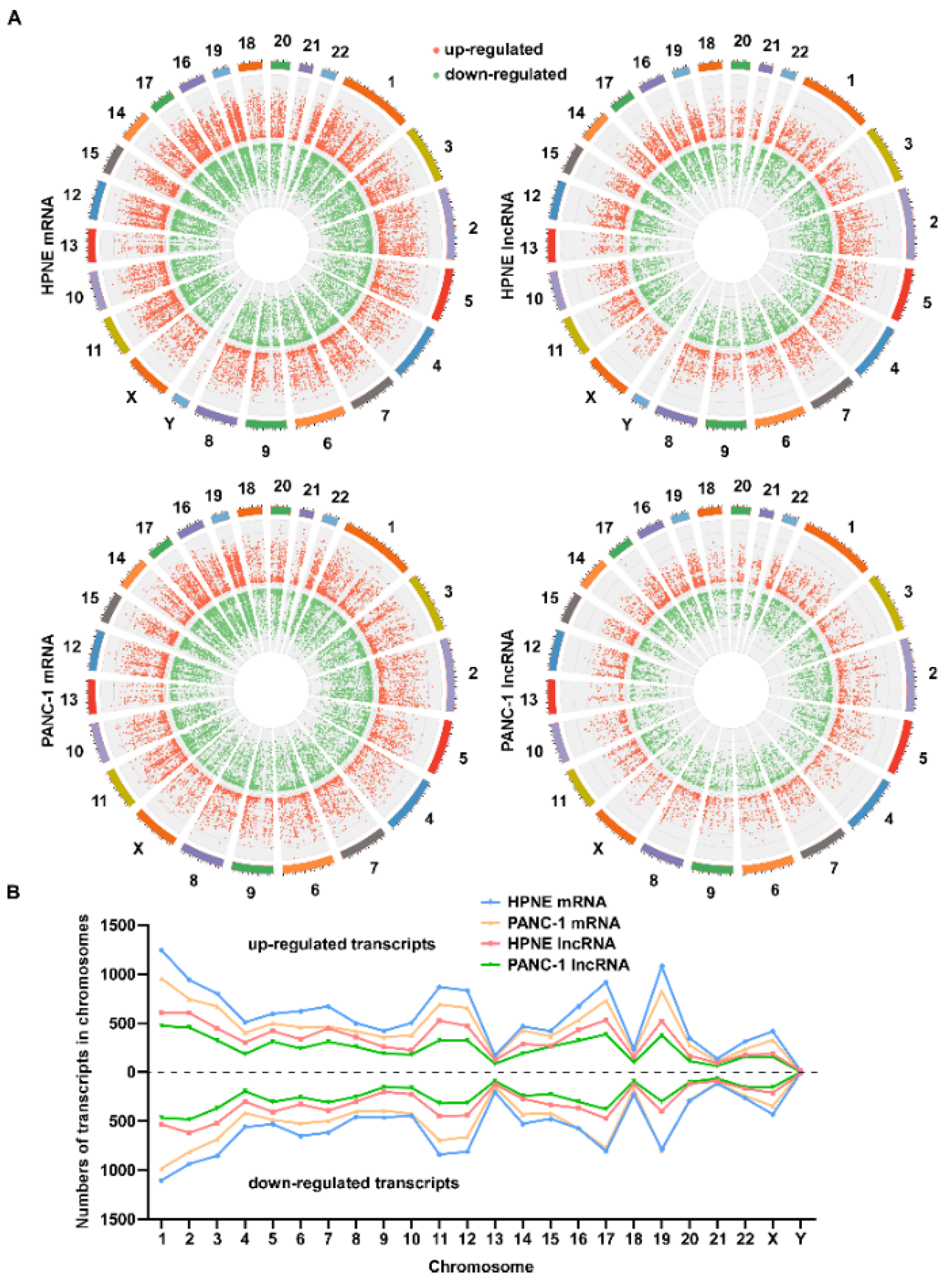
3.4. Chromosomal Distribution of Differentially Expressed Transcripts Induced by H2O2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, J.; Hu, Y.; Lu, W.; Pelicano, H.; Huang, P. Novel action of paclitaxel against cancer cells: Bystander effect mediated by reactive oxygen species. Cancer Res. 2007, 67, 3512–3517. [Google Scholar] [CrossRef]
- Conour, J.E.; Graham, W.V.; Gaskins, H.R. A combined in vitro/bioinformatic investigation of redox regulatory mechanisms governing cell cycle progression. Physiol. Genom. 2004, 18, 196–205. [Google Scholar] [CrossRef][Green Version]
- Trachootham, D.; Alexandre, J.; Huang, P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat. Rev. Drug Discov. 2009, 8, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef]
- Schippers, J.H.; Nguyen, H.M.; Lu, D.; Schmidt, R.; Mueller-Roeber, B. ROS homeostasis during development: An evolutionary conserved strategy. Cell. Mol. Life Sci. 2012, 69, 3245–3257. [Google Scholar] [CrossRef]
- Le Gal, K.; Schmidt, E.E.; Sayin, V.I. Cellular Redox Homeostasis. Antioxidants 2021, 10, 1377. [Google Scholar] [CrossRef] [PubMed]
- Trachootham, D.; Lu, W.; Ogasawara, M.A.; Nilsa, R.V.; Huang, P. Redox regulation of cell survival. Antioxid. Redox Signal. 2008, 10, 1343–1374. [Google Scholar] [CrossRef]
- Kohlgruber, S.; Upadhye, A.; Dyballa-Rukes, N.; McNamara, C.A.; Altschmied, J. Regulation of Transcription Factors by Reactive Oxygen Species and Nitric Oxide in Vascular Physiology and Pathology. Antioxid. Redox Signal. 2017, 26, 679–699. [Google Scholar] [CrossRef]
- Turpaev, K.T. Reactive oxygen species and regulation of gene expression. Biochemistry 2002, 67, 281–292. [Google Scholar]
- Morgan, M.J.; Liu, Z.G. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef]
- Lingappan, K. NF-kappaB in Oxidative Stress. Curr. Opin. Toxicol. 2018, 7, 81–86. [Google Scholar] [CrossRef]
- Dhar, A.; Young, M.R.; Colburn, N.H. The role of AP-1, NF-kappaB and ROS/NOS in skin carcinogenesis: The JB6 model is predictive. Mol. Cell. Biochem. 2002, 234–235, 185–193. [Google Scholar] [CrossRef]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef]
- Kovac, S.; Angelova, P.R.; Holmstrom, K.M.; Zhang, Y.; Dinkova-Kostova, A.T.; Abramov, A.Y. Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochim. Biophys. Acta 2015, 1850, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Hagen, T. Oxygen versus Reactive Oxygen in the Regulation of HIF-1alpha: The Balance Tips. Biochem. Res. Int. 2012, 2012, 436981. [Google Scholar] [CrossRef] [PubMed]
- Qutub, A.A.; Popel, A.S. Reactive oxygen species regulate hypoxia-inducible factor 1alpha differentially in cancer and ischemia. Mol. Cell. Biol. 2008, 28, 5106–5119. [Google Scholar] [CrossRef]
- Konno, T.; Melo, E.P.; Chambers, J.E.; Avezov, E. Intracellular Sources of ROS/H2O2 in Health and Neurodegeneration: Spotlight on Endoplasmic Reticulum. Cells 2021, 10, 233. [Google Scholar] [CrossRef]
- Bienert, G.P.; Schjoerring, J.K.; Jahn, T.P. Membrane transport of hydrogen peroxide. Biochim. Biophys. Acta 2006, 1758, 994–1003. [Google Scholar] [CrossRef]
- Dayalan Naidu, S.; Kostov, R.V.; Dinkova-Kostova, A.T. Transcription factors Hsf1 and Nrf2 engage in crosstalk for cytoprotection. Trends Pharmacol. Sci. 2015, 36, 6–14. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Li, C.H.; Chen, Y. Insight Into the Role of Long Noncoding RNA in Cancer Development and Progression. Int. Rev. Cell Mol. Biol. 2016, 326, 33–65. [Google Scholar]
- Wang, X.; Shen, C.; Zhu, J.; Shen, G.; Li, Z.; Dong, J. Long Noncoding RNAs in the Regulation of Oxidative Stress. Oxidative Med. Cell. Longev. 2019, 2019, 1318795. [Google Scholar] [CrossRef]
- Connerty, P.; Lock, R.B.; de Bock, C.E. Long Non-coding RNAs: Major Regulators of Cell Stress in Cancer. Front. Oncol. 2020, 10, 285. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Shoorei, H.; Taheri, M. Non-coding RNAs are involved in the response to oxidative stress. Biomed. Pharmacother. 2020, 127, 110228. [Google Scholar] [CrossRef] [PubMed]
- Glorieux, C.; Xia, X.; He, Y.Q.; Hu, Y.; Cremer, K.; Robert, A.; Liu, J.; Wang, F.; Ling, J.; Chiao, P.J.; et al. Regulation of PD-L1 expression in K-ras-driven cancers through ROS-mediated FGFR1 signaling. Redox Biol. 2021, 38, 101780. [Google Scholar] [CrossRef]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef]
- Lennicke, C.; Cocheme, H.M. Redox metabolism: ROS as specific molecular regulators of cell signaling and function. Mol. Cell 2021, 81, 3691–3707. [Google Scholar] [CrossRef]
- Picazo, C.; Molin, M. Impact of Hydrogen Peroxide on Protein Synthesis in Yeast. Antioxidants 2021, 10, 952. [Google Scholar] [CrossRef]
- Cox, L.S.; Lane, D.P. Tumour suppressors, kinases and clamps: How p53 regulates the cell cycle in response to DNA damage. Bioessays 1995, 17, 501–508. [Google Scholar] [CrossRef]
- Principe, D.R.; Timbers, K.E.; Atia, L.G.; Koch, R.M.; Rana, A. TGFbeta Signaling in the Pancreatic Tumor Microenvironment. Cancers 2021, 13, 5086. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ye, W.; Zhang, R.; He, X.; Wang, J.; Liu, Y.; Wang, Y.; Yang, X.; Zhang, Y.; Gan, W. The Role of TGF-beta Signaling Pathways in Cancer and Its Potential as a Therapeutic Target. Evid.-Based Complement. Altern. Med. 2021, 2021, 6675208. [Google Scholar]
- Liu, S.; Chen, S.; Zeng, J. TGFbeta signaling: A complex role in tumorigenesis (Review). Mol. Med. Rep. 2018, 17, 699–704. [Google Scholar] [PubMed]
- Thomas, S.J.; Snowden, J.A.; Zeidler, M.P.; Danson, S.J. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer 2015, 113, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Kietzmann, T.; Petry, A.; Shvetsova, A.; Gerhold, J.M.; Görlach, A. The epigenetic landscape related to reactive oxygen species formation in the cardiovascular system. Br. J. Pharmacol. 2017, 174, 1533–1554. [Google Scholar] [CrossRef] [PubMed]
- Kreuz, S.; Fischle, W. Oxidative stress signaling to chromatin in health and disease. Epigenomics 2016, 8, 843–862. [Google Scholar] [CrossRef] [PubMed]


| Term | Description | Log P |
|---|---|---|
| R-HSA-1640170 | Cell cycle | −44.7433 |
| R-HSA-8953854 | Metabolism of RNA | −44.3152 |
| R-HSA-194315 | Signaling by Rho GTPases | −23.5974 |
| GO:1903047 | Mitotic cell-cycle process | −22.8387 |
| WP3888 | VEGFA–VEGFR2 signaling pathway | −22.3273 |
| GO:0051603 | Proteolysis involved in cellular protein catabolic process | −21.2771 |
| R-HSA-157118 | Signaling by NOTCH | −19.9154 |
| R-HSA-3700989 | Transcriptional regulation by TP53 | −19.7004 |
| R-HSA-5663202 | Diseases of signal transduction by growth factor receptors and second messengers | −19.0701 |
| R-HSA-1280215 | Cytokine signaling in immune system | −19.0064 |
| GO:0051301 | Cell division | −17.9398 |
| R-HSA-109581 | Apoptosis | −17.2555 |
| R-HSA-69275 | G2/M transition | −16.8881 |
| GO:0097190 | Apoptotic signaling pathway | −16.0895 |
| GO:0034976 | Response to endoplasmic reticulum stress | −15.357 |
| R-HSA-72203 | Processing of capped intron-containing pre-mRNA | −14.5354 |
| R-HSA-109582 | Hemostasis | −13.6556 |
| GO:0097435 | Supramolecular fiber organization | −13.653 |
| GO:1903827 | Regulation of cellular protein localization | −13.3734 |
| ko05203 | Viral carcinogenesis | −13.26 |
| R-HSA-1640170 | Cell cycle | −44.7433 |
| R-HSA-8953854 | Metabolism of RNA | −44.3152 |
| Group | Total | Elements |
|---|---|---|
| lncRNA-up + lncRNA-down + mRNA-up + mRNA-down | 6 | STAT3, RELA, TP53, NFKB1, SP1, ESR1 |
| lncRNA-up + mRNA-up + mRNA-down | 3 | MYCN, MYC, JUN |
| lncRNA-down + mRNA-up + mRNA-down | 2 | TFAP2A, AR |
| lncRNA-up + mRNA-up | 7 | ETS1, YY1, TP73, RB1, NFE2L2, KLF6, ATM |
| lncRNA-up + mRNA-down | 1 | HIF1A |
| lncRNA-down + mRNA-up | 2 | BRCA1, SP3 |
| lncRNA-down + mRNA-down | 2 | SATB1, SOX6 |
| mRNA-up + mRNA-down | 11 | YBX1, ATF2, ATF3, HDAC1, EGR1, SIRT1, E2F1, NR3C1, KLF4, HDAC2, CREB1 |
| lncRNA-up | 1 | EP300 |
| lncRNA-down | 1 | NFIC |
| mRNA-up | 21 | CDX2, TCF3, PPARG, SREBF1, ATF4, WT1, STAT1, MYBL2, JUND, POU2F1, POU5F1, BCL6, STAT5A, RARA, ETV6, NFYA, CEBPB, USF2, USF1, SMAD7, NRF1 |
| mRNA-down | 11 | TWIST2, E2F3, TWIST1, SRF, NF1, MSC, HMGA1, IRF1, EZH2, ETS2, HDAC3 |
| lncRNA-up + lncRNA-down + mRNA-up + mRNA-down | 6 | STAT3, RELA, TP53, NFKB1, SP1, ESR1 |
| lncRNA-up + mRNA-up + mRNA-down | 3 | MYCN, MYC, JUN |
| lncRNA-down + mRNA-up + mRNA-down | 2 | TFAP2A, AR |
| Group | Total | Elements |
|---|---|---|
| lncRNA-up + lncRNA-down + mRNA-up + mRNA-down | 1 | SP1 |
| lncRNA-up + lncRNA-down + mRNA-up | 1 | MYC |
| lncRNA-up + mRNA-up | 1 | E2F1 |
| lncRNA-down + mRNA-down | 1 | MYCN |
| mRNA-up + mRNA-down | 2 | JUN, EZH2 |
| lncRNA-up | 4 | RELA, NFKB1, SP3, EP300 |
| mRNA-up | 6 | DDIT3, HIF1A, TP53, SIRT1, AR, ESR1 |
| mRNA-down | 5 | TWIST2, YY1, TWIST1, STAT1, PPARG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Qiu, Y.; Xiang, R.; Huang, P. Characterization of H2O2-Induced Alterations in Global Transcription of mRNA and lncRNA. Antioxidants 2022, 11, 495. https://doi.org/10.3390/antiox11030495
Liu S, Qiu Y, Xiang R, Huang P. Characterization of H2O2-Induced Alterations in Global Transcription of mRNA and lncRNA. Antioxidants. 2022; 11(3):495. https://doi.org/10.3390/antiox11030495
Chicago/Turabian StyleLiu, Shihua, Ya Qiu, Rong Xiang, and Peng Huang. 2022. "Characterization of H2O2-Induced Alterations in Global Transcription of mRNA and lncRNA" Antioxidants 11, no. 3: 495. https://doi.org/10.3390/antiox11030495
APA StyleLiu, S., Qiu, Y., Xiang, R., & Huang, P. (2022). Characterization of H2O2-Induced Alterations in Global Transcription of mRNA and lncRNA. Antioxidants, 11(3), 495. https://doi.org/10.3390/antiox11030495







