Transcriptome and Metabolome Analysis of a Late-Senescent Vegetable Soybean during Seed Development Provides New Insights into Degradation of Chlorophyll
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sample Collection
2.2. Measurement of Chl Content and Fluorescence Intensity
2.3. Pod Moisture Content, Seed Moisture Content, and Seed Dry Weight Determination
2.4. Soluble Sugars, Starch, and Antioxidant Enzyme Activity Determinations
2.5. Stranded-RNA-Seq Library Construction and Sequencing
2.6. Read Mapping and Quantifying
2.7. Annotation of Differentially Expressed Genes
2.8. Real-Time Quantitative PCR Analysis
2.9. Metabolic Analysis
2.10. Data Availability
2.11. Statistic Analysis
3. Results
3.1. ZX12 Seeds Showed Higher Chl Content and Lower Chl Degradation Rate When Compared with ZX4
3.2. Physiological Character of ZX4 and ZX12 Seeds at Different Stages of Maturity
3.3. Transcriptome Analysis of ZX4 and ZX12 Seeds at Different Stages of Maturity Revealed Differential Expression of Chl Metabolism and Photosynthesis Pathway-Related Genes
3.4. Metabolome Analysis of ZX4 and ZX12 Seeds at Different Stages of Maturity Identified a Differential Abundance of Flavonoids
3.5. Correlation Analysis of Differential Metabolites and DEGs in the Flavonoid and Anthocyanin Pathways
4. Discussion
4.1. Unique Delayed Senescence Characteristics Suggest a New Regulatory Mechanism
4.2. Genes Relating to Chl Metabolism and Photosynthesis Are Involved in Regulating the Late-Senescent Phenotype
4.3. Flavonoids Inhibit Chlorophyll Degradation by Scavenging Oxygen Free Radicals
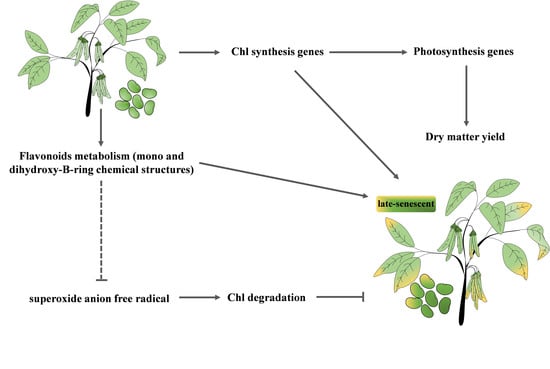
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Williams Ii, M.M. Phenomorphological Characterization of Vegetable Soybean Germplasm Lines for Commercial Production. Crop Sci. 2015, 55, 1274–1279. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; Chin, K.L.; Qi, Y. Vegetable soybean: Seed composition and production research. Ital. J. Agron. 2017, 12, 276–282. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.C. Food Safety and Contract Edamame: The Geopolitics of the Vegetable Trade in East Asia. Geogr. Rev. 2018, 108, 274–295. [Google Scholar] [CrossRef]
- Zhang, L.; Kyei-Boahen, S. Growth and Yield of Vegetable Soybean (Edamame) in Mississippi. Horttechnology 2007, 17, 26–31. [Google Scholar] [CrossRef] [Green Version]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, M.; Sciarappa, W.J.; Simon, J.E. LC/UV/ESI-MS Analysis of Isoflavones in Edamame and Tofu Soybeans. J. Agric. Food Chem. 2004, 52, 2763. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.L.; Katuuramu, D.N. Comparison of seed fatty and amino acids in edamame dried using two oven-drying methods and mature soybeans. J. Sci. Food Agric. 2021, 101, 1515–1522. [Google Scholar] [CrossRef]
- Xu, Y.; Cartier, A.; Kibet, D.; Jordan, K.; Hakala, I.; Davis, S.; Sismour, E.; Kering, M.; Rutto, L. Physical and nutritional properties of edamame seeds as influenced by stage of development. J. Food Meas. Charact. 2016, 10, 193–200. [Google Scholar] [CrossRef]
- Zhang, J.; Fengler, K.A.; Van Hemert, J.L.; Gupta, R.; Mongar, N.; Sun, J.; Allen, W.B.; Wang, Y.; Weers, B.; Mo, H. Identification and characterization of a novel stay-green QTL that increases yield in maize. Plant Biotechnol. J. 2019, 17, 2272–2285. [Google Scholar] [CrossRef]
- Thomas, H.; Howarth, C. Five ways to stay green. J. Exp. Bot. 2000, 51, 329–337. [Google Scholar] [CrossRef]
- Yang, K.; An, J.P.; Li, C.Y.; Shen, X.N.; Liu, Y.J.; Wang, D.R.; Ji, X.L.; Hao, Y.J.; You, C.X. The apple C2H2-type zinc finger transcription factor MdZAT10 positively regulates JA-induced leaf senescence by interacting with MdBT2. Hortic. Res. 2021, 8, 159. [Google Scholar] [CrossRef] [PubMed]
- Akimoto, K.; Kuroda, S. Quality of Green Soybeans Packaged in Perforated PE/PP Film. J. Jpn. Soc. Hortic. Sci. 1981, 50, 100–107. [Google Scholar] [CrossRef] [Green Version]
- Ergo, V.V.; Veas, R.E.; Vega, C.R.C.; Lascano, R.; Carrera, C.S. Leaf photosynthesis and senescence in heated and droughted field-grown soybean with contrasting seed protein concentration. Plant Physiol. Biochem. 2021, 166, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Himelblau, E.; Amasino, R.M. Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J. Plant Physiol. 2001, 158, 1317–1323. [Google Scholar] [CrossRef]
- Wang, H.; Schippers, J.H.M. The Role and Regulation of Autophagy and the Proteasome During Aging and Senescence in Plants. Genes 2019, 10, 267. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, S. Molecular regulation of leaf senescence. Curr. Opin. Plant Biol. 2003, 6, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, P.; Culetic, A.; Boschian, L.; Krupinska, K. Plant senescence and crop productivity. Plant Mol. Biol. 2013, 82, 603–622. [Google Scholar] [CrossRef]
- Li, X.; Ma, Y.; Li, P.; Zhang, L.; Wang, Y.; Zhang, R.; Wang, N. RNAi-mediated knocking-down ofrlpk2 gene retarded soybean leaf senescence. Chin. Sci. Bull. 2005, 50, 1218–1224. [Google Scholar] [CrossRef]
- Thomas, H. Senescence, ageing and death of the whole plant. New Phytol. 2013, 197, 696–711. [Google Scholar] [CrossRef]
- Smolikova, G.; Dolgikh, E.; Vikhnina, M.; Frolov, A.; Medvedev, S. Genetic and Hormonal Regulation of Chlorophyll Degradation during Maturation of Seeds with Green Embryos. Int. J. Mol. Sci. 2017, 18, 1993. [Google Scholar] [CrossRef]
- Nakajima, S.; Ito, H.; Tanaka, R.; Tanaka, A. Chlorophyll b Reductase Plays an Essential Role in Maturation and Storability of Arabidopsis Seeds. Plant Physiol. 2012, 160, 261–273. [Google Scholar] [CrossRef] [Green Version]
- Smolikova, G.; Laman, N.; Boriskevich, O. Role of Chlorophylls and Carotenoids in Seed Tolerance to Abiotic Stressors. Russ. J. Plant Physiol. 2011, 58, 965–973. [Google Scholar] [CrossRef]
- Ye, J.; Yang, W.; Li, Y.; Shiwen, W.; Yin, L.; Deng, X.P. Seed Pre-Soaking with Melatonin Improves Wheat Yield by Delaying Leaf Senescence and Promoting Root Development. Agronomy 2020, 10, 84. [Google Scholar] [CrossRef] [Green Version]
- Hörtensteiner, S. Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 2009, 14, 14–155. [Google Scholar] [CrossRef] [PubMed]
- Harbach, C.J.; Allen, T.W.; Bowen, C.R.; Davis, J.A.; Hill, C.B.; Leitman, M.; Leonard, B.R.; Mueller, D.S.; Padgett, G.B.; Phillips, X.A.; et al. Delayed Senescence in Soybean: Terminology, Research Update, and Survey Results from Growers. Plant Health Prog. 2016, 17, 76–83. [Google Scholar] [CrossRef] [Green Version]
- Kusaba, M.; Ito, H.; Morita, R.; Iida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M.; et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex ii and grana degradation during leaf senescence. Plant Cell 2007, 19, 1362–1375. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2008, 57, 120–131. [Google Scholar] [CrossRef]
- Meguro, M.; Ito, H.; Takabayashi, A.; Tanaka, R.; Tanaka, A. Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell 2011, 23, 3442–3453. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Kim, Y.S.; Yoo, S.C.; Hörtensteiner, S.; Paek, N.C. 7-Hydroxymethyl chlorophyll a reductase functions in metabolic channeling of chlorophyll breakdown intermediates during leaf senescence. Biochem. Biophys. Res. Commun. 2013, 430, 32–37. [Google Scholar] [CrossRef] [Green Version]
- Xie, Q.; Liang, Y.; Zhang, J.; Zheng, H.; Dong, G.; Qian, Q.; Zuo, J. Involvement of a Putative Bipartite Transit Peptide in Targeting Rice Pheophorbide a Oxygenase into Chloroplasts for Chlorophyll Degradation during Leaf Senescence. J. Genet. Genom. 2016, 43, 145–154. [Google Scholar] [CrossRef]
- Das, A.; Christ, B.; Hörtensteiner, S. Characterization of the pheophorbide a oxygenase/phyllobilin pathway of chlorophyll breakdown in grasses. Planta 2018, 248, 875–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schelbert, S.; Aubry, S.; Burla, B.; Agne, B.; Kessler, F.; Krupinska, K.; Hörtensteiner, S. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 2009, 21, 767–785. [Google Scholar] [CrossRef] [Green Version]
- Fang, C.; Li, C.; Li, W.; Wang, Z.; Zhou, Z.; Shen, Y.; Wu, M.; Wu, Y.; Li, G.; Kong, L.A.; et al. Concerted evolution of D1 and D2 to regulate chlorophyll degradation in soybean. Plant J. 2014, 77, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wu, S.; Li, N.; Gao, J.; Liu, S.; Zhu, S.; Li, Z.; Ren, G.; Kuai, B. Chemical induction of leaf senescence and powdery mildew resistance involves ethylene-mediated chlorophyll degradation and ROS metabolism in cucumber. Hortic. Res. 2022, 9, uhac101. [Google Scholar] [CrossRef] [PubMed]
- Niu, F.; Cui, X.; Zhao, P.; Sun, M.; Yang, B.; Deyholos, M.K.; Li, Y.; Zhao, X.; Jiang, Y.Q. WRKY42 transcription factor positively regulates leaf senescence through modulating SA and ROS synthesis in Arabidopsis thaliana. Plant J. 2020, 104, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Ott, A.; Yang, Y.; Bhattacharyya, M.; Horner, H.T.; Palmer, R.G.; Sandhu, D. Molecular Mapping of D1, D2 and ms5 Revealed Linkage between the Cotyledon Color Locus D2 and the Male-Sterile Locus ms5 in Soybean. Plants 2013, 2, 441–454. [Google Scholar] [CrossRef] [Green Version]
- Luquez, V.M.; Guiamét, J.J. Effects of the ‘Stay Green’ Genotype GGd1d1d2d2 on Leaf Gas Exchange, Dry Matter Accumulation and Seed Yield in Soybean (Glycine max L. Merr.). Ann. Bot. 2001, 87, 313–318. [Google Scholar] [CrossRef] [Green Version]
- Thomas, H.; Smart, C.M. Crops that Stay Green. Ann. Appl. Biol. 1993, 123, 193–219. [Google Scholar] [CrossRef]
- Guiamét, J.J.; Tyystjärvi, E.; Tyystjärvi, T.; John, I.; Kairavuo, M.; Pichersky, E.; Noodén, L.D. Photoinhibition and loss of photosystem II reaction centre proteins during senescence of soybean leaves. Enhancement of photoinhibition by the ‘stay-green’ mutation cytG. Physiol. Plant. 2002, 115, 468–478. [Google Scholar] [CrossRef] [Green Version]
- Myers, J.R.; Aljadi, M.; Brewer, L. The Importance of Cosmetic Stay-Green in Specialty Crops. In Plant Breeding Reviews; Goldman, I., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2018; Volume 42, pp. 219–256. [Google Scholar]
- Nakano, M.; Yamada, T.; Masuda, Y.; Sato, Y.; Kobayashi, H.; Ueda, H.; Morita, R.; Nishimura, M.; Kitamura, K.; Kusaba, M. A Green-Cotyledon/Stay-Green Mutant Exemplifies the Ancient Whole-Genome Duplications in Soybean. Plant Cell Physiol. 2014, 55, 1763–1771. [Google Scholar] [CrossRef]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuraba, Y.; Schelbert, S.; Park, S.Y.; Han, S.H.; Lee, B.D.; Andres, C.B.; Kessler, F.; Hortensteiner, S.; Paek, N.C. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 2012, 24, 507–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Gao, L.; Li, R.Z.; Wang, Y.; Liu, Y.Y.; Zhang, X.; Xie, H. High-throughput sequencing reveals the molecular mechanisms determining the stay-green characteristic in soybeans. J. Biosci. 2020, 45, 103. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. In Methods in Enzymology; Packer, L., Douce, R., Eds.; Academic Press: New York, NY, USA, 1987; Volume 148, pp. 350–382. [Google Scholar]
- Hou, G.; Littlejohn, S.S. Visualization of Green and Red Leaf Structures in Flowering Pear Pyrus Calleryana Using Integrated Microscopy. J. Plant Sci. 2014, 1, 28–32. [Google Scholar]
- Khonchaisri, R.; Sumonsiri, N.; Prommajak, T.; Rachtanapun, P.; Leksawasdi, N.; Techapun, C.; Taesuwan, S.; Halee, A.; Nunta, R.; Khemacheewakul, J. Optimization of Ultrasonic-Assisted Bioactive Compound Extraction from Green Soybean (Glycine max L.) and the Effect of Drying Methods and Storage Conditions on Procyanidin Extract. Foods 2022, 11, 1775. [Google Scholar] [CrossRef]
- Dubois, M.; Giles, K.; Rebers, P.; Smith, F. Colorimetric method for determination of sugar and related substances. Anal. Chem. 1955, 28, 350–356. [Google Scholar] [CrossRef]
- Xu, C.J.; Chen, W.J.; Chen, K.S.; Zhang, S.L. A simple method for determining the content of starch-iodine colorimety. Biotechnology 1998, 8, 41–43. [Google Scholar]
- Liu, C.H.; Di, Y.P. Analysis of RNA Sequencing Data Using CLC Genomics Workbench. In Molecular Toxicology Protocols; Keohavong, P., Singh, K.P., Gao, W., Eds.; Springer: New York, NY, USA, 2020; pp. 61–113. [Google Scholar]
- Boyle, E.I.; Weng, S.; Jeremy, G.; Jin, H.; David, B.; Michael, C.J.; Gavin, S. GO::TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004, 18, 3710–3715. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Hu, R.; Fan, C.; Hongyu, L.; Zhang, Q.; Fu, Y.F. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 2009, 10, 93. [Google Scholar] [CrossRef] [Green Version]
- Thomas, H.; Ougham, H. The stay-green trait. J. Exp. Bot. 2014, 65, 3889–3900. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.D.; Yan, Y.F.; Kim, M.Y.; Lee, S.H.; Lee, B.W. Population-specific quantitative trait loci mapping for functional stay-green trait in rice (Oryza sativa L.). Genome 2011, 54, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.C.; Cho, S.H.; Zhang, H.; Paik, H.C.; Paek, N.C. Quantitative trait loci associated with functional stay-green SNU-SG1 in rice. Mol. Cells 2007, 24, 83–94. [Google Scholar]
- Fu, J.D.; Yan, Y.F.; Lee, B.W. Physiological characteristics of a functional stay-green rice “SNU-SG1” during grain-filling period. J. Crop Sci. Biotechnol. 2009, 12, 47–52. [Google Scholar] [CrossRef]
- Ren, T.; Fan, T.; Chen, S.; Chen, Y.; Ou, X.; Jiang, Q.; Peng, W.; Ren, Z.; Tan, F.; Luo, P.; et al. Identification and validation of quantitative trait loci for the functional stay green trait in common wheat (Triticum aestivum L.) via high-density SNP-based genotyping. Theor. Appl. Genet. 2022, 135, 1429–1441. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhang, Y.; Huang, S.; Liu, Z.; Li, C.; Feng, H. Defect in Brnym1, a magnesium-dechelatase protein, causes a stay-green phenotype in an EMS-mutagenized Chinese cabbage (Brassica campestris L. ssp. pekinensis) line. Hortic. Res. 2020, 7, 1–11. [Google Scholar] [CrossRef]
- Wang, H.; Hou, L.; Wang, M.; Mao, P. Contribution of the pod wall to seed grain filling in alfalfa. Sci. Rep. 2016, 6, 26586. [Google Scholar] [CrossRef] [Green Version]
- Li, X.P.; Gan, R.; Li, P.L.; Ma, Y.Y.; Zhang, L.W.; Zhang, R.; Wang, Y.; Wang, N.N. Identification and functional characterization of a leucine-rich repeat receptor-like kinase gene that is involved in regulation of soybean leaf senescence. Plant Mol. Biol. 2006, 61, 829–844. [Google Scholar] [CrossRef]
- Wang, P.; Hou, S.Y.; Wen, H.W.; Wang, Q.Z.; Li, G.Q. Chlorophyll retention caused by STAY-GREEN (SGR) gene mutation enhances photosynthetic efficiency and yield in soybean hybrid Z1. Photosynthetica 2020, 59, 37–48. [Google Scholar] [CrossRef]
- Shalygo, N.; Czarnecki, O.; Peter, E.; Grimm, B. Expression of chlorophyll synthase is also involved in feedback-control of chlorophyll biosynthesis. Plant Mol. Biol. 2009, 71, 425–436. [Google Scholar] [CrossRef]
- Shah, A.A.; Yasin, N.A.; Mudassir, M.; Ramzan, M.; Hussain, I.; Siddiqui, M.H.; Ali, H.M.; Shabbir, Z.; Ali, A.; Ahmed, S.; et al. Iron oxide nanoparticles and selenium supplementation improve growth and photosynthesis by modulating antioxidant system and gene expression of chlorophyll synthase (CHLG) and protochlorophyllide oxidoreductase (POR) in arsenic-stressed Cucumis melo. Environ. Pollut. 2022, 307, 119413. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.M.; Söll, D. Antisense HEMA1 RNA Expression Inhibits Heme and Chlorophyll Biosynthesis in Arabidopsis. Plant Physiol. 2000, 122, 49–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paddock, T.; Mason, M.; Lima, D.; Armstrong, G. Arabidopsis protochlorophyllide oxidoreductase A (PORA) restores bulk chlorophyll synthesis and normal development to a porB porC double mutant. Plant Mol. Biol. 2009, 72, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.O.; Yerramsetty, P.; Zielinski, A.M.; Mure, C.M. Photosynthetic gene expression in higher plants. Photosynth. Res. 2013, 117, 91–120. [Google Scholar] [CrossRef]
- Mirkovic, T.; Ostroumov, E.E.; Anna, J.M.; van Grondelle, R.; Govindjee; Scholes, G.D. Light Absorption and Energy Transfer in the Antenna Complexes of Photosynthetic Organisms. Chem. Rev. 2017, 117, 249–293. [Google Scholar] [CrossRef]
- Ben-Shem, A.; Frolow, F.; Nelson, N. Crystal structure of plant photosystem I. Nature 2003, 426, 630–635. [Google Scholar] [CrossRef]
- Ludewig, F.; Sonnewald, U. High CO2-mediated down-regulation of photosynthetic gene transcripts is caused by accelerated leaf senescence rather than sugar accumulation. FEBS Lett. 2000, 479, 19–24. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, L.F. Plant Secondary Metabolites: From Molecular Biology to Health Products; Azooz, M.M., Ahmad, P., Eds.; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2015; pp. 263–299. [Google Scholar]
- Guo, Y.; Wang, T.; Fu, F.F.; El-Kassaby, Y.A.; Wang, G. Temporospatial Flavonoids Metabolism Variation in Ginkgo biloba Leaves. Front. Genet. 2020, 11, 589326. [Google Scholar] [CrossRef]
- Pietta, P.G. Flavonoids as Antioxidants. J. Nat. Prod. 2000, 63, 1035–1042. [Google Scholar] [CrossRef]
- Singh, P.; Arif, Y.; Bajguz, A.; Hayat, S. The role of quercetin in plants. Plant Physiol. Biochem. 2021, 166, 10–19. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, D.; Shen, Y.; Ni, Z.; Wang, Q.; Lei, Z.; Xu, N.; Deng, Q.; Lin, L.; Wang, J.; Lv, X.; et al. Exogenous Melatonin Application Delays Senescence of Kiwifruit Leaves by Regulating the Antioxidant Capacity and Biosynthesis of Flavonoids. Front. Plant Sci. 2018, 9, 426. [Google Scholar] [CrossRef] [PubMed]
- Procházková, D.; Wilhelmová, N. Leaf senescence and activities of the antioxidant enzymes. Biol. Plant. 2007, 51, 401–406. [Google Scholar] [CrossRef]
- Pal, L.; Kar, R.K. Role of Reactive Oxygen Species in Cotyledon Senescence During Early Seedling Stage of Mung Bean [Vigna radiata (L.) Wilczek]. J. Plant Growth Regul. 2019, 38, 315–324. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef]
- Inoue, K. Emerging roles of the chloroplast outer envelope membrane. Trends Plant Sci. 2011, 16, 550–557. [Google Scholar] [CrossRef]
- Agati, G.; Matteini, P.; Goti, A.; Tattini, M. Chloroplast-located flavonoids can scavenge singlet oxygen. New Phytol. 2007, 174, 77–89. [Google Scholar] [CrossRef]
- Moellering, E.R.; Muthan, B.; Benning, C. Freezing Tolerance in Plants Requires Lipid Remodeling at the Outer Chloroplast Membrane. Science 2010, 330, 226–228. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Chen, G.; Li, X.; Fu, X.; Li, S.; Tao, X.; Chen, Z.-H.; Xu, S. Transcriptome and Metabolome Analysis of a Late-Senescent Vegetable Soybean during Seed Development Provides New Insights into Degradation of Chlorophyll. Antioxidants 2022, 11, 2480. https://doi.org/10.3390/antiox11122480
Wang J, Chen G, Li X, Fu X, Li S, Tao X, Chen Z-H, Xu S. Transcriptome and Metabolome Analysis of a Late-Senescent Vegetable Soybean during Seed Development Provides New Insights into Degradation of Chlorophyll. Antioxidants. 2022; 11(12):2480. https://doi.org/10.3390/antiox11122480
Chicago/Turabian StyleWang, Jian, Guang Chen, Xuetong Li, Xujun Fu, Sujuan Li, Xiaoyuan Tao, Zhong-Hua Chen, and Shengchun Xu. 2022. "Transcriptome and Metabolome Analysis of a Late-Senescent Vegetable Soybean during Seed Development Provides New Insights into Degradation of Chlorophyll" Antioxidants 11, no. 12: 2480. https://doi.org/10.3390/antiox11122480