Identification of Copy Number Aberrations in Breast Cancer Subtypes Using Persistence Topology
Abstract
:1. Introduction
2. Experimental Section
2.1. Simulation Data
2.2. The Horlings Dataset
2.3. Detection of Focal Copy Number Aberrations Using TAaCGH
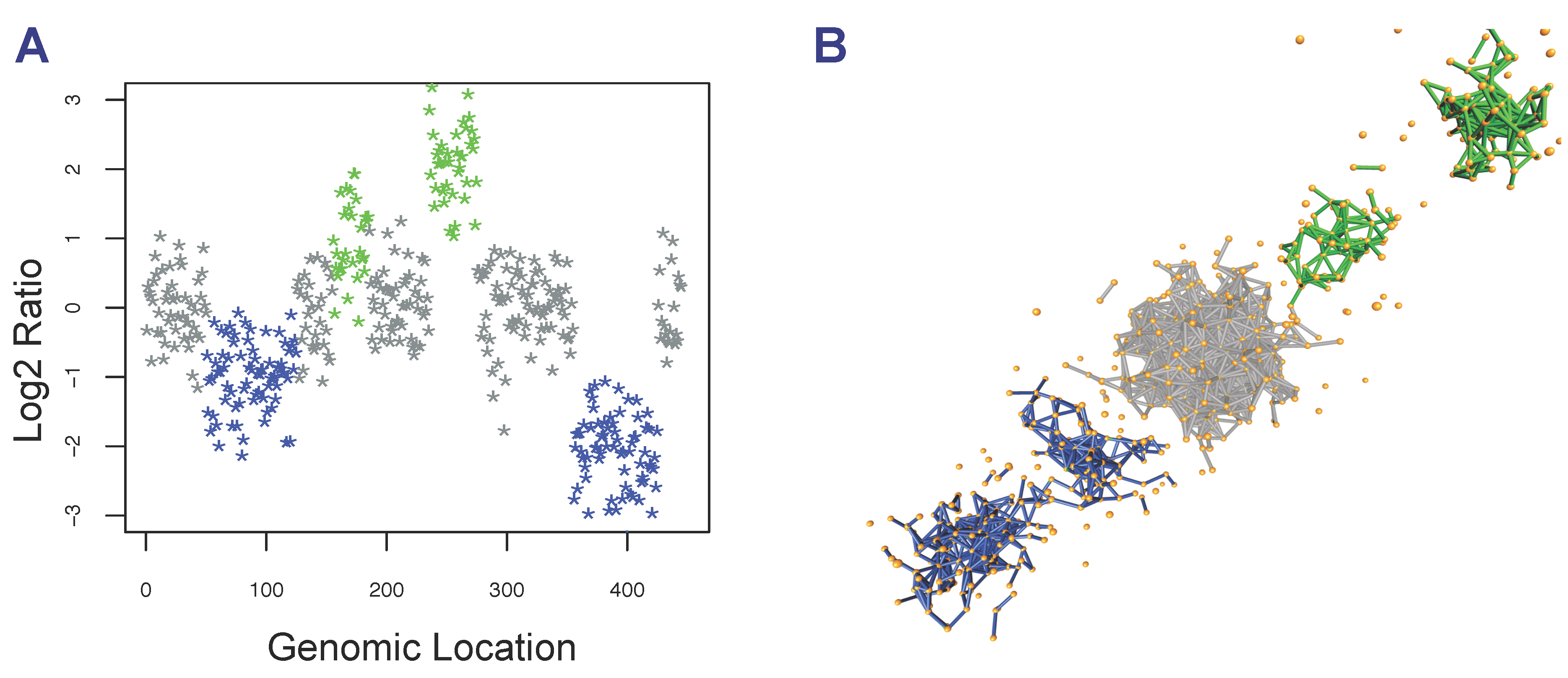
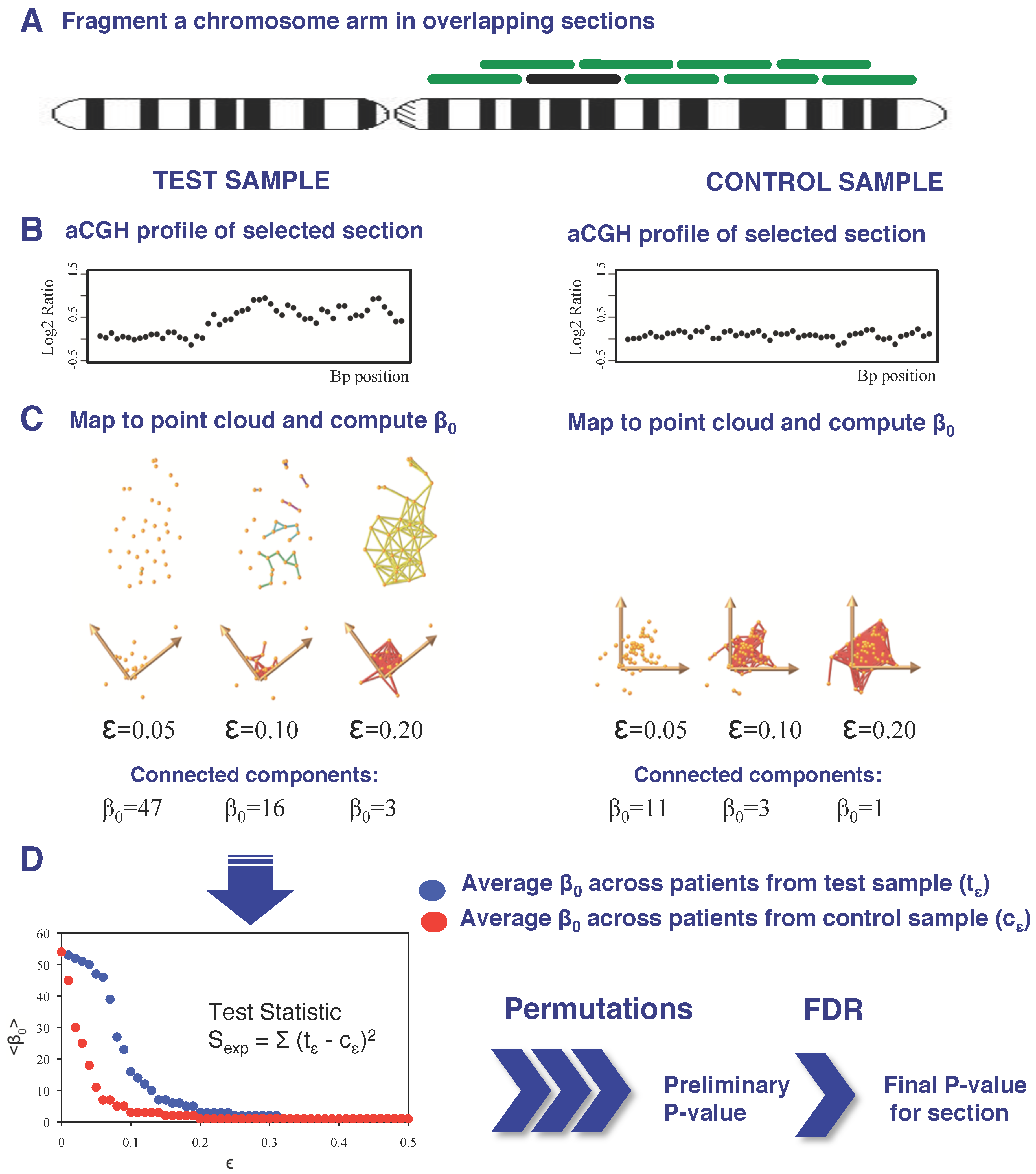
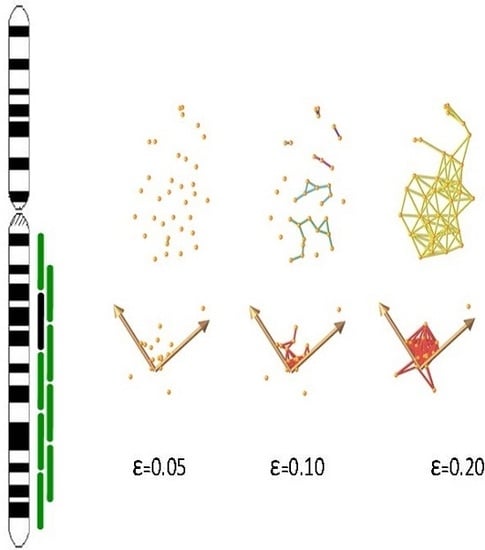
2.4. Determining Significance of Specific Clones
2.5. Detection of Full-Length Arm/Chromosome Section Aberrations
2.6. Validation of the Experimental Results
3. Results and Discussion
3.1. Simulation Results
3.1.1. Window Size
| Dimensions | 2 | 5 | 10 | 15 | 20 | 35 | 50 |
|---|---|---|---|---|---|---|---|
| 2 | 1 | 0.89 | 0.92 | 0.95 | 0.93 | 0.93 | 0.89 |
| 5 | 1 | 0.91 | 0.92 | 0.92 | 0.91 | 0.85 | |
| 10 | 1 | 0.95 | 0.94 | 0.95 | 0.84 | ||
| 15 | 1 | 0.98 | 0.97 | 0.93 | |||
| 20 | 1 | 0.99 | 0.91 | ||||
| 35 | 1 | 0.93 |
3.1.2. Sensitivity and Specificity of TaACGH
3.1.3. Size of the Chromosome Section
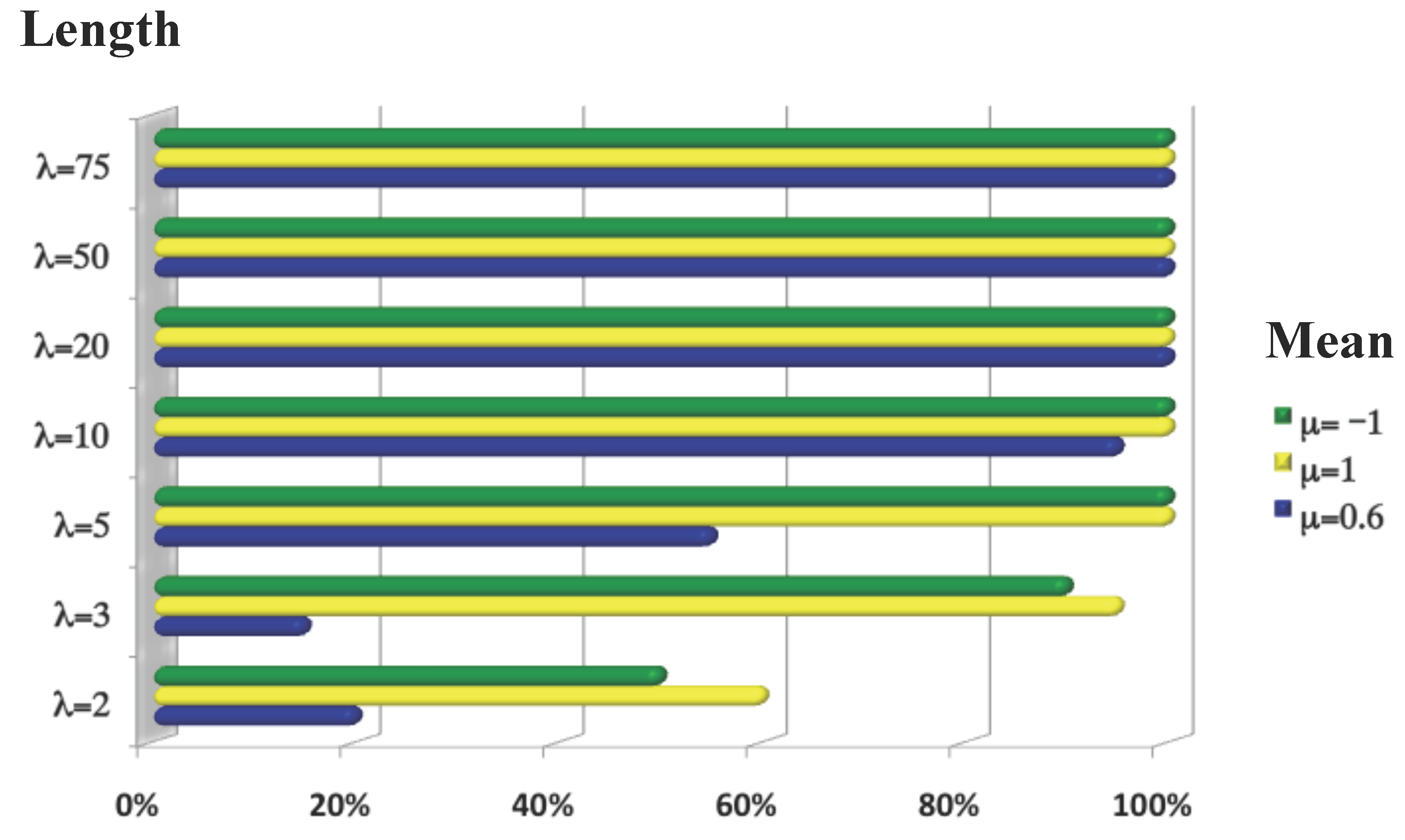
3.1.4. Performance of TAaCGH when Both Control and Test Population Have Overlapping Aberrations
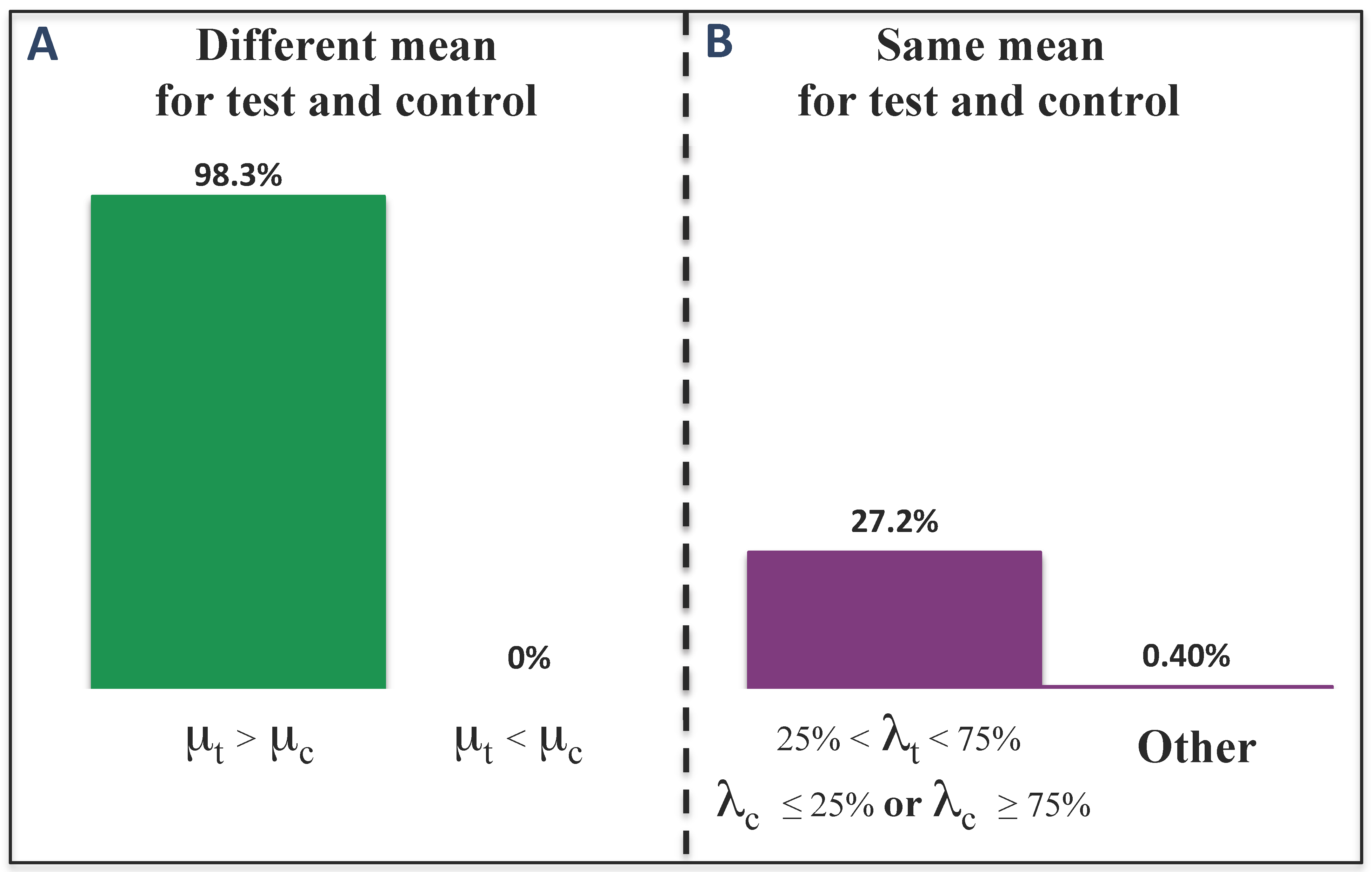
3.2. Results for Breast Cancer Subtypes
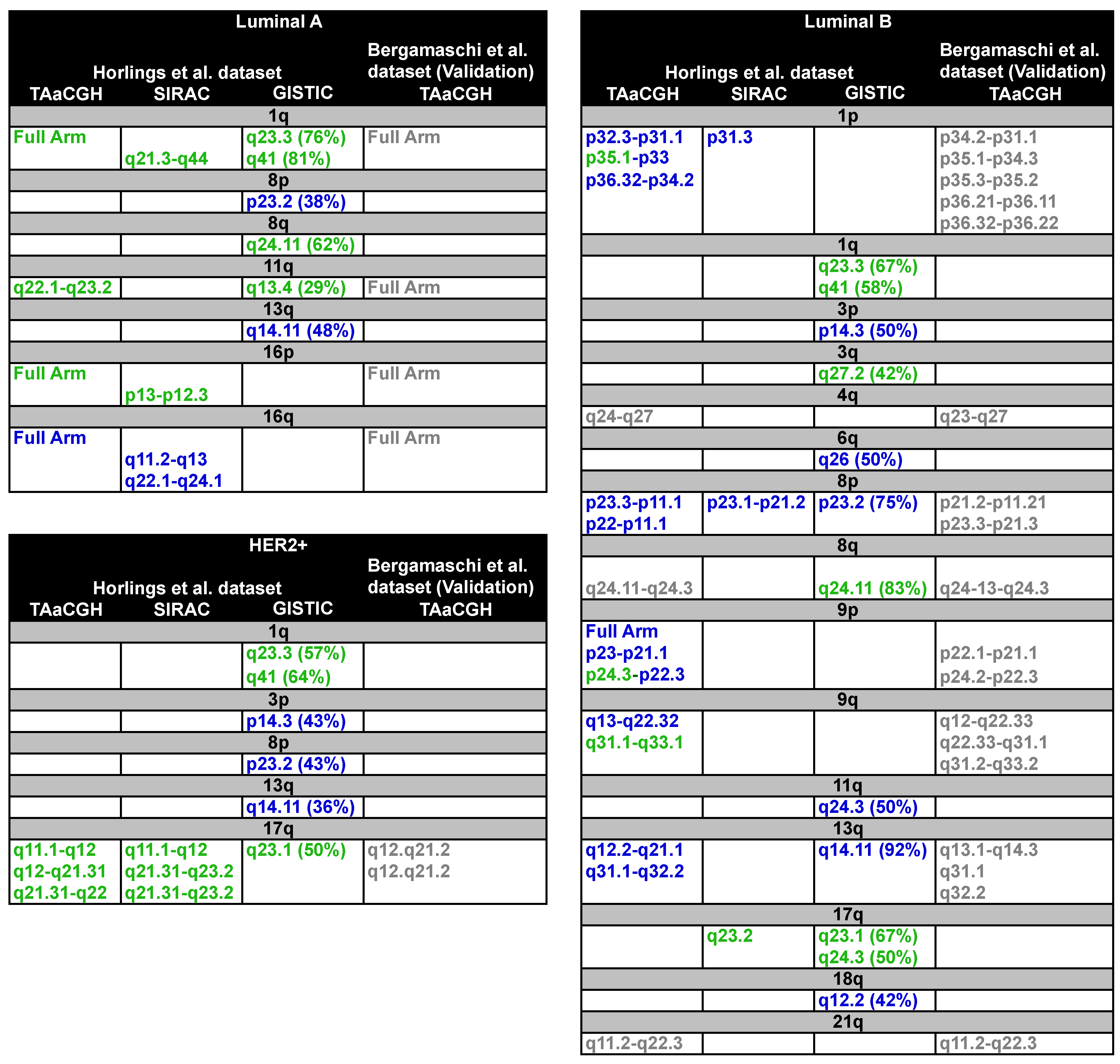

3.2.1. Analysis of Luminal Subtypes


| Chromosome Arm | Cytoband | Location of Aberration | Gain/Loss | Location of Neighboring Clones |
|---|---|---|---|---|
| 1p | 36.32 | 3,225,674 | loss | 3,225,674 to 4,577,827 |
| 36.22 | 10,154,043 | loss | 9,161,350 to 11,561,620 | |
| 36.22 | 11,561,620 | loss | 11,064,731 to 11,844,141 | |
| 36.22 | 12,429,632 | loss | 11,844,141 to 14,639,539 | |
| 36.21 | 15,553,913 | loss | 14,639,539 to 17,268,504 | |
| 36.12 | 23,338,991 | loss | 22,032,838 to 24,480,408 | |
| 36.11 | 27,703,779 | loss | 27,359,936 to 27,856,736 | |
| 34.3 | 35,105,342 | gain | 34,380,537 to 36,832,678 | |
| 34.3 | 38,492,287 | loss | 38,278,515 to 39,536,339 | |
| 33 | 50,571,477 | loss | 49,145,731 to 50,823,002 | |
| 31.33 to 31.1 | 65,599,782 to 70,103,164 | loss | 64,435,137 to 70,406,779 | |
| 31.1 | 76,869,240 | loss | 75,463,114 to 78,005,143 | |
| 9p | 24.1 | 6,004,718 | gain | 4,922,574 to 6,576,990 |
| 23 | 9,668,611 | loss | 8,409,615 to 9,943,073 | |
| 22.1 | 18,590,957 | loss | 17,850,221 to 19,321,518 | |
| 21.3 | 23,387,562 | loss | 22,490,595 to 24,101,721 | |
| 9q | 21.11 | 71,549,399 | loss | 71,129,855 to 72,258,752 |
| 21.31 | 83,762,927 | loss | 82,953,907 to 84,780,238 | |
| 22.2 | 93,147,096 | loss | 92,847,611 to 94,131,899 | |
| 31.1 | 106,545,018 | gain | 98,981,704 to 107,489,797 | |
| 13q | 12.2 to 31.1 | 32,170,305 to 41,470,434 | loss | 31,017,797 to 51,431,527 |
| 31.1 | 79,057,929 | loss | 56,818,886 to 81,814,181 | |
| 31.1 | 83,138,436 to 83,695,803 | loss | 81,974,786 to 84,869,575 | |
| 31.1 | 84,869,575 to 87,444,574 | loss | 83,695,803 to 88,048,738 |
3.2.2. Analysis of the ERBB2/HER2/NEU (HER2+)-Enriched Subtype

3.2.3. Results for the Basal-Like Subtype
- Chromosome arm 1q: We found the region 1q23.1 to 31.1 to be aberrant, and GISTIC confirmed it to be an amplification. This region was large enough that the changes were also detected in the study using the center of masses. This region of the genome was previously reported in other studies [31,32,36].
- Chromosome arm 3q: We found an amplification of the whole arm, while GISTIC found region 3q27.2. The gain of 3q is characteristic of BRCA1 deficiency in sporadic tumors, as well as in hereditary tumors (e.g., [71]).
- Chromosome arm 6q: Three sections out of nine were found amplified in chromosome arm 6q. These three regions expanded cytobands 6q24.1 to q27 and contain the estrogen receptor gene ERS1 located at 6q25.2. This finding was also detected in our study by the center of masses [69].
- Chromosome arm 12p: The region 12p13.3 was found to be amplified using our topological analysis, and the entire arm was also detected by the displacement of the center of masses. GISTIC detected a downstream region 12p33 to be amplified.
- Chromosome arm 13q: Two main sections were found aberrant in the chromosome arm 13q. The first one expanding 13q12.2 to q31.2 and the second 13q31.2 to q34. We were unable to identify whether 13q12.2 to q31.2 was an amplification or a deletion; however, GISTIC identified a deletion in 13q14.11 in 81% of the patients. Additionally TAaCGH found an amplification in 13q31.2 to q34. Amplifications in chromosome 13q have been found in multiple subtypes [36] and more specifically in cytokeratin 14 (CK14) positive tumors, 25% of which are basal-like carcinomas [72].
- Chromosome arm 18q: A section extending 18q11.1 to q21.33 was significant, but TAaCGH was unable to identify whether it was an amplification or a deletion, suggesting an heterogeneous combinations of amplifications and deletions in the region across subtypes. GISTIC, on the other hand, identified a deletion in 18q12.2.
| Chromosome Arm | Cytoband | Location of Aberration | Gain/Loss | Location of Neighboring Clones |
|---|---|---|---|---|
| 1p | 36.21 | 14,639,539 | loss | 12,429,632 to 15,553,913 |
| 35.1 to 32.3 | 34,380,537 to 53,737,606 | gain | 33,102,443 to 54,031,005 | |
| 32.3 | 55,875,826 | loss | 55,270,329 to 56,824,097 | |
| 32.2-31.3 | 58,882,124 to 62,359,674 | gain | 57,684,846 to 63,423,938 | |
| 31.1 | 78,005,143 | gain | 76,869,240 to 82,056,781 | |
| 22.1 | 93,996,738 | loss | 92,518,227 to 94,792,570 | |
| 21.2 | 99,899,192 | gain | 98,909,737 to 99,899,246 | |
| 21.1 | 101,303,797 | loss | 99,899,246 to 101,449,113 | |
| 21.2 | 101,449,113 | gain | 101,303,797 to 102,594,767 | |
| 13.3 | 107,585,795 to 108,529,492 | gain | 106,959,397 to 110,015,588 | |
| 13.1 | 117,424,118 | gain | 116,780,010 to 118,589,386 | |
| 2p | 14 to 13.3 | 64,625,779 to 71,023,979 | gain | 63,342,684 to 71,251,890 |
| 14q | 13.2 | 33,766,983 to 36,544,890 | loss | 33,310,161 to 37,941,646 |
| 21.1 | 37,941,646 | loss | 36,544,890 to 46,896,032 | |
| 21.2 | 44,219,870 | loss | 42,928,208 to 45,707,124 | |
| 24.3 | 76,584,595 | loss | 76,141,892 to 77,583,617 | |
| 31.1 | 80,271,909 to 83,099,727 | loss | 78,389,382 to 83,987,892 | |
| 31.2 | 84,784,824 | loss | 83,987,892 to 85,099,070 | |
| 31.3 | 87,007,447 to 87,344,554 | loss | 85,099,070 to 87,765,087 | |
| 31.3 to 32.12 | 89,750,111 to 92,321,034 | loss | 88,420,711 to 93,495,784 | |
| 32.13 to 32.2 | 94,877,177 to 97,996,975 | loss | 93,495,784 to 95,662,483 | |

- Chromosome arm . The section containing Xp22.33 to 11.21, which contained all clones in the array, was found significant in [72].
4. Conclusions
5. Software
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Garcia, M.A.; Geyer, F.C.; Lacroix-Triki, M.; Marchió, C.; Reis-Filho, J.S. Breast cancer precursors revisited: molecular features and progression pathways. Histopathology 2010, 57, 171–192. [Google Scholar] [CrossRef] [PubMed]
- Kwei, K.A.; Kung, Y.; Salari, K.; Holcomb, I.N.; Pollack, J.R. Genomic instability in breast cancer: Pathogenesis and clinical implications. Mol. Oncol. 2010, 4, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.W. Our changing view of the genomic landscape of cancer. J. Pathol. 2010, 220, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, S.F.; Gruel, N.; Chapeaublanc, E.; Lescure, A.; Jones, T.; Reyal, F.; Vincent-Salomon, A.; Raynal, V.; Pierron, G.; Perez, F.; et al. A siRNA screen identifies RAD21, EIF3H, CHRAC1 and TANC2 as driver genes within the 8q23, 8q24.3 and 17q23 amplicons in breast cancer with effects on cell growth, survival and transformation. Carcinogenesis 2014, 35, 670–682. [Google Scholar] [PubMed]
- Wang, E. Understanding genomic alterations in cancer genomes using an integrative network approach. Cancer Lett. 2013, 340, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Climent, J.; Dimitrow, P.; Fridlyand, J.; Palacios, J.; Siebert, R.; Albertson, D.G.; Gray, J.W.; Pinkel, D.; Lluch, A.; Martinez-Climent, J.A. Deletion of chromosome 11q predicts response to anthracycline-based chemotherapy in early breast cancer. Cancer Res. 2007, 67, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Climent, J.; Garcia, J.; Mao, J.; Arsuaga, J.; Perez-Losada, J. Characterization of breast cancer by array comparative genomic hybridization. This paper is one of a selection of papers published in this Special Issue, entitled 28th International West Coast Chromatin and Chromosome Conference, and has undergone the Journal’s usual peer review process. Biochem. Cell Biol. 2007, 85, 497–508. [Google Scholar]
- Desmedt, C.; Voet, T.; Sotiriou, C.; Campbell, P.J. Next generation sequencing in breast cancer: First take home messages. Curr. Opin. Oncol. 2012, 24, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.A.; Li, J.; Doig, K.; Fellowes, A.; Wong, S.Q. Studying Cancer Genomics Through Next-Generation DNA Sequencing and Bioinformatics. In Clinical Bioinformatics; Springer: New York, NY, USA, 2014; pp. 83–98. [Google Scholar]
- Meyerson, M.; Gabriel, S.; Getz, G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 2010, 11, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Pinkel, D.; Albertson, D.G. Array comparative genomic hybridization and its applications in cancer. Nat. Genet. 2005, 37, S11–S17. [Google Scholar] [CrossRef] [PubMed]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bignell, G.R.; Greenman, C.D.; Davies, H.; Butler, A.P.; Edkins, S.; Andrews, J.M.; Buck, G.; Chen, L.; Beare, D.; Latimer, C.; et al. Signatures of mutation and selection in the cancer genome. Nature 2010, 463, 893–898. [Google Scholar] [CrossRef] [PubMed]
- Boyle, J.; Kreisberg, R.; Bressler, R.; Killcoyne, S. Methods for visual mining of genomic and proteomic data atlases. BMC Bioinform. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wendl, M.C.; McMichael, J.F.; Raphael, B.J. Expanding the computational toolbox for mining cancer genomes. Nat. Rev. Genet. 2014, 15, 556–570. [Google Scholar] [CrossRef] [PubMed]
- Fridlyand, J.; Snijders, A.M.; Pinkel, D.; Albertson, D.G.; Jain, A.N. Hidden Markov models approach to the analysis of array CGH data. J. Multivar. Anal. 2004, 90, 132–153. [Google Scholar] [CrossRef]
- Hupé, P.; Stransky, N.; Thiery, J.P.; Radvanyi, F.; Barillot, E. Analysis of array CGH data: From signal ratio to gain and loss of DNA regions. Bioinformatics 2004, 20, 3413–3422. [Google Scholar] [CrossRef] [PubMed]
- Klijn, C.; Holstege, H.; de Ridder, J.; Liu, X.; Reinders, M.; Jonkers, J.; Wessels, L. Identification of cancer genes using a statistical framework for multiexperiment analysis of nondiscretized array CGH data. Nucleic Acids Res. 2008, 36. [Google Scholar] [CrossRef]
- De Ronde, J.J.; Klijn, C.; Velds, A.; Holstege, H.; Reinders, M.J.; Jonkers, J.; Wessels, L.F. KC-SMARTR: An R package for detection of statistically significant aberrations in multi-experiment aCGH data. BMC Res. Notes 2010, 3. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.; Horlings, H.M.; van de Vijver, M.J.; van Beers, E.H.; Nederlof, P.M.; Wessels, L.F.; Reinders, M.J. SIRAC: Supervised Identification of Regions of Aberration in aCGH datasets. BMC Bioinform. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- De Woskin, D.; Climent, J.; Cruz-White, I.; Vazquez, M.; Park, C.; Arsuaga, J. Applications of computational homology to the analysis of treatment response in breast cancer patients. Topol. Appl. 2010, 157, 157–164. [Google Scholar] [CrossRef]
- Perea, J.A.; Harer, J. Sliding windows and persistence: An application of topological methods to signal analysis. Found. Comput. Math. 2013, 15, 799–838. [Google Scholar] [CrossRef]
- Edelsbrunner, H.; Harer, J. Persistent homology-a survey. Contemp. Math. 2008, 453, 257–282. [Google Scholar]
- Zomorodian, A.J. Topology for computing; Cambridge University Press: Cambridge, UK, 2005; Volume 16. [Google Scholar]
- Hsu, L.; Self, S.G.; Grove, D.; Randolph, T.; Wang, K.; Delrow, J.J.; Loo, L.; Porter, P. Denoising array-based comparative genomic hybridization data using wavelets. Biostatistics 2005, 6, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.; van de Rijn, M.; Jeffrey, S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Sørlie, T.; Tibshirani, R.; Parker, J.; Hastie, T.; Marron, J.; Nobel, A.; Deng, S.; Johnsen, H.; Pesich, R.; Geisler, S.; et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA 2003, 100, 8418–8423. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Børresen-Dale, A.L. Systems biology and genomics of breast cancer. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Shiu, K.K.; Natrajan, R.; Geyer, F.C.; Ashworth, A.; Reis-Filho, J.S. DNA amplifications in breast cancer: Genotypic-phenotypic correlations. Future Oncol. 2010, 6, 967–984. [Google Scholar] [CrossRef] [PubMed]
- Adélaïde, J.; Finetti, P.; Bekhouche, I.; Repellini, L.; Geneix, J.; Sircoulomb, F.; Charafe-Jauffret, E.; Cervera, N.; Desplans, J.; Parzy, D.; et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007, 67, 11565–11575. [Google Scholar] [CrossRef] [PubMed]
- Bergamaschi, A.; Kim, Y.H.; Wang, P.; Sørlie, T.; Hernandez-Boussard, T.; Lonning, P.E.; Tibshirani, R.; Børresen-Dale, A.L.; Pollack, J.R. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer 2006, 45, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Horlings, H.M.; Lai, C.; Nuyten, D.S.; Halfwerk, H.; Kristel, P.; van Beers, E.; Joosse, S.A.; Klijn, C.; Nederlof, P.M.; Reinders, M.J.; et al. Integration of DNA copy number alterations and prognostic gene expression signatures in breast cancer patients. Clin. Cancer Res. 2010, 16, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, G.; Staaf, J.; Vallon-Christersson, J.; Ringnér, M.; Holm, K.; Hegardt, C.; Gunnarsson, H.; Fagerholm, R.; Strand, C.; Agnarsson, B.A.; et al. Research article Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Res. 2010, 12. [Google Scholar] [CrossRef] [PubMed]
- Loo, L.W.; Grove, D.I.; Williams, E.M.; Neal, C.L.; Cousens, L.A.; Schubert, E.L.; Holcomb, I.N.; Massa, H.F.; Glogovac, J.; Li, C.I.; et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004, 64, 8541–8549. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar]
- Bilal, E.; Vassallo, K.; Toppmeyer, D.; Barnard, N.; Rye, I.H.; Almendro, V.; Russnes, H.; Børresen-Dale, A.L.; Levine, A.J.; Bhanot, G.; et al. Amplified loci on chromosomes 8 and 17 predict early relapse in ER-positive breast cancers. PLoS ONE 2012, e38575. [Google Scholar] [CrossRef] [PubMed]
- Climent, J.; Martinez-Climent, J.A.; Blesa, D.; Garcia-Barchino, M.J.; Saez, R.; Sánchez-Izquierdo, D.; Azagra, P.; Lluch, A.; Garcia-Conde, J. Genomic loss of 18p predicts an adverse clinical outcome in patients with high-risk breast cancer. Clin. Cancer Res. 2002, 8, 3863–3869. [Google Scholar] [PubMed]
- Han, W.; Han, M.R.; Kang, J.J.; Bae, J.Y.; Lee, J.H.; Bae, Y.J.; Lee, J.E.; Shin, H.J.; Hwang, K.T.; Hwang, S.E.; et al. Genomic alterations identified by array comparative genomic hybridization as prognostic markers in tamoxifen-treated estrogen receptor-positive breast cancer. BMC Cancer 2006, 6. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.T.; Han, W.; Cho, J.; Lee, J.W.; Ko, E.; Kim, E.K.; Jung, S.Y.; Jeong, E.M.; Bae, J.Y.; Kang, J.J.; et al. Genomic copy number alterations as predictive markers of systemic recurrence in breast cancer. Int. J. Cancer 2008, 123, 1807–1815. [Google Scholar] [CrossRef] [PubMed]
- McDonald, S.L.; Stevenson, D.A.; Moir, S.E.; Hutcheon, A.W.; Haites, N.E.; Heys, S.D.; Schofield, A.C. Genomic changes identified by comparative genomic hybridisation in docetaxel-resistant breast cancer cell lines. Eur. J. Cancer 2005, 41, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Rouzier, R.; Perou, C.M.; Symmans, W.F.; Ibrahim, N.; Cristofanilli, M.; Anderson, K.; Hess, K.R.; Stec, J.; Ayers, M.; Wagner, P.; et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin. Cancer Res. 2005, 11, 5678–5685. [Google Scholar] [CrossRef] [PubMed]
- Seute, A.; Sinn, H.P.; Schlenk, R.F.; Emig, R.; Wallwiener, D.; Grischke, E.M.; Hohaus, S.; Döhner, H.; Haas, R.; Bentz, M. Clinical relevance of genomic aberrations in homogeneously treated high-risk stage II/III breast cancer patients. Int. J. Cancer 2001, 93, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Urquidi, V.; Goodison, S. Derivation of molecular signatures for breast cancer recurrence prediction using a two-way validation approach. Breast Cancer Res. Treat. 2010, 119, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.; Lambros, M.B.; Horlings, H.M.; Pearson, A.; Sharpe, R.; Natrajan, R.; Geyer, F.C.; van Kouwenhove, M.; Kreike, B.; Mackay, A.; et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene 2010, 29, 2013–2023. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.R.; Johnson, M.D.; Kucherlapati, R.; Park, P.J. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics 2005, 21, 3763–3770. [Google Scholar] [CrossRef] [PubMed]
- Troyanskaya, O.; Cantor, M.; Sherlock, G.; Brown, P.; Hastie, T.; Tibshirani, R.; Botstein, D.; Altman, R.B. Missing value estimation methods for DNA microarrays. Bioinformatics 2001, 17, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Arsuaga, J.; Baas, N.A.; DeWoskin, D.; Mizuno, H.; Pankov, A.; Park, C. Topological analysis of gene expression arrays identifies high risk molecular subtypes in breast cancer. Appl. Algebra Eng. Commun. Comput. 2012, 23, 3–15. [Google Scholar] [CrossRef]
- Sexton, H.; Vejdemo-Johansson, M. jPlex, 2008. Available online: http://comptop.stanford.edu/programs/jplex/ (accessed on 1 March 2015).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995, 57, 289–300. [Google Scholar]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Chin, K.; DeVries, S.; Fridlyand, J.; Spellman, P.T.; Roydasgupta, R.; Kuo, W.L.; Lapuk, A.; Neve, R.M.; Qian, Z.; Ryder, T.; et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 2006, 10, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Olshen, A.B.; Venkatraman, E.; Lucito, R.; Wigler, M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004, 5, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Ades, F.; Zardavas, D.; Bozovic-Spasojevic, I.; Pugliano, L.; Fumagalli, D.; de Azambuja, E.; Viale, G.; Sotiriou, C.; Piccart, M. Luminal B breast cancer: Molecular characterization, clinical management, and future perspectives. J. Clin. Oncol. 2014, 32, 2794–2803. [Google Scholar] [CrossRef] [PubMed]
- Fridlyand, J.; Snijders, A.M.; Ylstra, B.; Li, H.; Olshen, A.; Segraves, R.; Dairkee, S.; Tokuyasu, T.; Ljung, B.M.; Jain, A.N.; et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer 2006, 6. [Google Scholar] [CrossRef] [PubMed]
- Natrajan, R.; Lambros, M.B.; Geyer, F.C.; Marchio, C.; Tan, D.S.; Vatcheva, R.; Shiu, K.K.; Hungermann, D.; Rodriguez-Pinilla, S.M.; Palacios, J.; et al. Loss of 16q in high grade breast cancer is associated with estrogen receptor status: Evidence for progression in tumors with a luminal phenotype? Genes Chromosomes Cancer 2009, 48, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Bièche, I.; Khodja, A.; Lidereau, R. Deletion mapping of chromosomal region 1p32-pter in primary breast cancer. Genes Chromosomes Cancer 1999, 24, 255–263. [Google Scholar] [CrossRef]
- Chin, S.; Wang, Y.; Thorne, N.; Teschendorff, A.; Pinder, S.; Vias, M.; Naderi, A.; Roberts, I.; Barbosa-Morais, N.; Garcia, M.; et al. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene 2007, 26, 1959–1970. [Google Scholar] [CrossRef] [PubMed]
- Ray, M.E.; Yang, Z.Q.; Albertson, D.; Kleer, C.G.; Washburn, J.G.; Macoska, J.A.; Ethier, S.P. Genomic and expression analysis of the 8p11–12 amplicon in human breast cancer cell lines. Cancer Res. 2004, 64, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Streicher, K.; Yang, Z.; Draghici, S.; Ethier, S. Transforming function of the LSM1 oncogene in human breast cancers with the 8p11–12 amplicon. Oncogene 2007, 26, 2104–2114. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Q.; Albertson, D.; Ethier, S.P. Genomic organization of the 8p11–p12 amplicon in three breast cancer cell lines. Cancer Genet. Cytogenet. 2004, 155, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Q.; Streicher, K.L.; Ray, M.E.; Abrams, J.; Ethier, S.P. Multiple interacting oncogenes on the 8p11–p12 amplicon in human breast cancer. Cancer Res. 2006, 66, 11632–11643. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, S.; Liu, G.; Dombkowski, A.; Abrams, J.; Martin-Trevino, R.; Wicha, M.; Ethier, S.; Yang, Z. Identification and functional analysis of 9p24 amplified genes in human breast cancer. Oncogene 2012, 31, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Carlson, R.W.; Moench, S.J.; Hammond, M.; Perez, E.A.; Burstein, H.J.; Allred, D.C.; Vogel, C.L.; Goldstein, L.J.; Somlo, G.; Gradishar, W.J. HER2 testing in breast cancer: NCCN Task Force report and recommendations. J. Natl. Compr. Cancer Netw. 2006, 4, S1–S22. [Google Scholar]
- Wolff, A.C.; Hammond, M.E.H.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 2013, 31, 3997–4013. [Google Scholar] [CrossRef] [PubMed]
- Staaf, J.; Jonsson, G.; Ringner, M.; Vallon-Christersson, J.; Grabau, D.; Arason, A.; Gunnarsson, H.; Agnarsson, B.A.; Malmstrom, P.O.; Johannsson, O.T.; Loman, N.; Barkardottir, R.B.; Borg, A. High-resolution genomic and expression analyses of copy number alterations in HER2-amplified breast cancer. Breast Cancer Res. 2010, 12. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Niu, Y.; Wang, X.; Wei, L.; Lu, S. Genetic changes at specific stages of breast cancer progression detected by comparative genomic hybridization. J. Mol. Med. 2009, 87, 145–152. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, M.M.C.; de Oliveira, S.F.V.; Lima, R.S.; de Andrade Urban, C.; Cavalli, L.R.; Ribeiro, E.M.d.S.F.; Cavalli, I.J. Differential loss of heterozygosity profile on chromosome 3p in ductal and lobular breast carcinomas. Hum. Pathol. 2012, 43, 1661–1667. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Toher, J.; Morgan, M.; Davison, J.; Tannenbaum, S.; Claffey, K. Genomic differences between estrogen receptor (ER)-positive and ER-negative human breast carcinoma identified by single nucleotide polymorphism array comparative genome hybridization analysis. Cancer 2011, 117, 2024–2034. [Google Scholar] [CrossRef] [PubMed]
- Qian, P.; Banerjee, A.; Wu, Z.S.; Zhang, X.; Wang, H.; Pandey, V.; Zhang, W.J.; Lv, X.F.; Tan, S.; Lobie, P.E.; et al. Loss of SNAIL regulated miR-128-2 on chromosome 3p22.3 targets multiple stem cell factors to promote transformation of mammary epithelial cells. Cancer Res. 2012, 72, 6036–6050. [Google Scholar] [PubMed]
- Joosse, S.A.; Brandwijk, K.I.; Mulder, L.; Wesseling, J.; Hannemann, J.; Nederlof, P.M. Genomic signature of BRCA1 deficiency in sporadic basal-like breast tumors. Genes Chromosomes Cancer 2011, 50, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.; Ford, E.; Gillett, C.; Ryder, K.; Merrett, S.; Reis-Filho, J.S.; Fulford, L.G.; Hanby, A.; Lakhani, S.R. Molecular cytogenetic identification of subgroups of grade III invasive ductal breast carcinomas with different clinical outcomes. Clin. Cancer Res. 2004, 10, 5988–5997. [Google Scholar] [CrossRef] [PubMed]
- Vincent-Salomon, A.; Gruel, N.; Lucchesi, C.; MacGrogan, G.; Dendale, R.; Sigal-Zafrani, B.; Longy, M.; Raynal, V.; Pierron, G.; de Mascarel, I.; et al. Identification of typical medullary breast carcinoma as a genomic sub-group of basal-like carcinomas, a heterogeneous new molecular entity. Breast Cancer Res. 2007, 9. [Google Scholar] [CrossRef] [PubMed]
- Nicolau, M.; Levine, A.J.; Carlsson, G. Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival. Proc. Natl. Acad. Sci. 2011, 108, 7265–7270. [Google Scholar] [CrossRef] [PubMed]
- Steiner, M.A. Classification of breast cancer subtypes using signaling pathways and persistent homology. Master’s thesis, San Francisco State University, 2015. [Google Scholar]
- Forbes, S.; Bhamra, G.; Bamford, S.; Dawson, E.; Kok, C.; Clements, J.; Menzies, A.; Teague, J.; Futreal, P.; Stratton, M. The catalogue of somatic mutations in cancer (COSMIC). Curr. Protoc. Hum. Genet. 2008, 10–11. [Google Scholar]
- Jönsson, G.; Naylor, T.L.; Vallon-Christersson, J.; Staaf, J.; Huang, J.; Ward, M.R.; Greshock, J.D.; Luts, L.; Olsson, H.; Rahman, N.; et al. Distinct genomic profiles in hereditary breast tumors identified by array-based comparative genomic hybridization. Cancer Res. 2005, 65, 7612–7621. [Google Scholar] [PubMed]
- Dziegeil, P.; Owczarek, T.; Plazuk, E.; Gomułkiewicz, A.; Majchrzak, M.; Podhorska-Okołów, M.; Driouch, K.; Lidereau, R.; Ugorski, M. Ceramide galactosyltransferase (UGT8) is a molecular marker of breast cancer malignancy and lung metastases. Br. J. Cancer 2010, 103, 524–531. [Google Scholar]
- Ruckhäberle, E.; Karn, T.; Rody, A.; Hanker, L.; Gätje, R.; Metzler, D.; Holtrich, U.; Kaufmann, M. Gene expression of ceramide kinase, galactosyl ceramide synthase and ganglioside GD3 synthase is associated with prognosis in breast cancer. J. Cancer Res. Clin. Oncol. 2009, 135, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Opresko, P.L.; Calvo, J.P.; von Kobbe, C. Role for the Werner syndrome protein in the promotion of tumor cell growth. Mech. Ageing Dev. 2007, 128, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Pole, J.; Courtay-Cahen, C.; Garcia, M.; Blood, K.; Cooke, S.; Alsop, A.; Tse, D.; Caldas, C.; Edwards, P. High-resolution analysis of chromosome rearrangements on 8p in breast, colon and pancreatic cancer reveals a complex pattern of loss, gain and translocation. Oncogene 2006, 25, 5693–5706. [Google Scholar] [CrossRef] [PubMed]
- Suhasini, A.N.; Brosh, R.M. Disease-causing missense mutations in human DNA helicase disorders. Mutation Res. 2013, 752, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Turner-Ivey, B.; Guest, S.T.; Irish, J.C.; Kappler, C.S.; Garrett-Mayer, E.; Wilson, R.C.; Ethier, S.P. KAT6A, a Chromatin Modifier from the 8p11–p12 Amplicon is a Candidate Oncogene in Luminal Breast Cancer. Neoplasia 2014, 16, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, B.; Fabius, A.W.; Wu, W.H.; Pedraza, A.; Brennan, C.W.; Schultz, N.; Pitter, K.L.; Bromberg, J.F.; Huse, J.T.; Holland, E.C.; et al. Loss of the tyrosine phosphatase PTPRD leads to aberrant STAT3 activation and promotes gliomagenesis. Proc. Natl. Acad. Sci. 2014, 111, 8149–8154. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, B.; White, J.R.; Wu, W.H.; Chan, T.A. Deletion of Ptprd and Cdkn2a cooperate to accelerate tumorigenesis. Oncotarget 2014, 5, 6976–6982. [Google Scholar] [PubMed]
- An, H.X.; Claas, A.; Savelyeva, L.; Seitz, S.; Schlag, P.; Scherneck, S.; Schwab, M. Two regions of deletion in 9p23–24 in sporadic breast cancer. Cancer Res. 1999, 59, 3941–3943. [Google Scholar] [PubMed]
- Wernicke, M.; Roitman, P.; Manfre, D.; Stern, R. Breast cancer and the stromal factor. The prometastatic healing proces hypothesis. Medicina (B Aires) 2011, 71, 15–21. [Google Scholar]
- Cavalcante, R. Using homology and networks to locate copy number aberrations associated to recurrence in breast cancer. Master’s thesis, San Francisco State University, 2012. [Google Scholar]
- Rebouh, M. Exploring topological methods to study genomic imbalance in breast cancer. Master’s thesis, San Francisco State University, 2012. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arsuaga, J.; Borrman, T.; Cavalcante, R.; Gonzalez, G.; Park, C. Identification of Copy Number Aberrations in Breast Cancer Subtypes Using Persistence Topology. Microarrays 2015, 4, 339-369. https://doi.org/10.3390/microarrays4030339
Arsuaga J, Borrman T, Cavalcante R, Gonzalez G, Park C. Identification of Copy Number Aberrations in Breast Cancer Subtypes Using Persistence Topology. Microarrays. 2015; 4(3):339-369. https://doi.org/10.3390/microarrays4030339
Chicago/Turabian StyleArsuaga, Javier, Tyler Borrman, Raymond Cavalcante, Georgina Gonzalez, and Catherine Park. 2015. "Identification of Copy Number Aberrations in Breast Cancer Subtypes Using Persistence Topology" Microarrays 4, no. 3: 339-369. https://doi.org/10.3390/microarrays4030339