Comparative Analyses of MicroRNA Microarrays during Cardiogenesis: Functional Perspectives
Abstract
:1. Introduction
2. Experimental Section
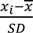 ). MicroRNA expression data in ventricular development [14], also Z-scored, was identically processed as describe above, but with an initial k-nearest neighbor(KNN) imputation of the densitometry values <1 using the KNN algorithm implemented in the Bioconductor impute package [15] with default parameters. Hierarchical clustering (Euclidean distance and complete linkage), an unsupervised way of grouping samples based only on their gene expression similarities, was carried out using TM4 software suite [17].
). MicroRNA expression data in ventricular development [14], also Z-scored, was identically processed as describe above, but with an initial k-nearest neighbor(KNN) imputation of the densitometry values <1 using the KNN algorithm implemented in the Bioconductor impute package [15] with default parameters. Hierarchical clustering (Euclidean distance and complete linkage), an unsupervised way of grouping samples based only on their gene expression similarities, was carried out using TM4 software suite [17].3. MicroRNAs in Cardiovascular Development
| Differentially expressed microRNAs | Cardio-vascular role | Validated targets (cardiovascular role) | Cardiac formation | iPS cardio-myogenesis | Cardiac aging | References |
|---|---|---|---|---|---|---|
| let-7a | nras, kras, hmga2 | up | up | down | [24] | |
| let-7b | up | up | na | |||
| let-7c | up | up | na | |||
| let-7d | up | up | na | |||
| let-7i | up | up | na | |||
| miR-15b | miR-15 | dmtf1, c22orf5, bcl-2, Chek1 | up | down | na | [18,24] |
| miR-17 | miR-17 | rbl2-p130, ncoa3, e2f1, adkcnna-p21, fog2 | up | na | na | [24,25,26] |
| miR-23b | miR-23 | pou4f2, hes1, has2 | up | up | na | [24,27] |
| miR-24 | miR-24 | notch1,mapk14, kiaa0152, dhfr, cdkn2a-p16, alk4, gata2, pak4, bcl2 | up | up | na | [24,28,29] |
| miR-25 | miR-25 | na | up | down | na | |
| miR-26a | up | up | na | |||
| miR-30a | miR-30 | ctgf, znf294, wnt5a, uap1, tnfrsf10b, tnfaip2, tmem87a, tmem59, tmem41b, tmed7, tmed3, tmed2, tmed10, tmco1, tloc1, ticam2, them4, sypl1, stx7, strn, slc9a3r2, slc9a3r2, slc7a11, slc7a1, slc4a7, slo4a10, slc38a2, slc38a1, slc12a4, sec23a, rpcd1, rbms1, rad23b, rab27b, ptrh1, ptprk, ptgfrn, prpf40a, ppp3r1, ppp3ca, ppp2r4, pgm1, ptprk, ptgfrn, prpf40a, ppp3r1, ppp3ca, ppp2r4, pgm1, pex11b, pafah1b2, pfha2, nufip2, nucb1, nt5e, nt5c3, npr3, np, ncl, napg, myo10, mpdu1, mllt11, mllt1, met, mbnl1, mat2a, lrrc8c, lmnb2, krthb5, kdelc2, jun, itga2, ifrd1, idh1, hnrpm, gpd2, gnai2, gilnt7, galnt1, fxr2, frg1, f2, elmod2, dock7, cpne8, chd1 | up | na | na | [30,31,32] |
| miR-30d | up | up | na | |||
| miR-93 | miR-93 | na | up | down | na | |
| miR-99a | up | up | up | |||
| miR-99b | up | up | na | |||
| miR-103 | up | down | na | |||
| miR-106a | up | down | down | |||
| miR-122a | miR-122 | trpr6, ndrg3, cd320, bckdk, aldoa, cck-8, caspase-3 | up | na | na | [24,33] |
| miR-125a | miR-125 | Lin28, erbb2, erbb3, zfp385, tor2a, rhebl1, ppt2, mkk7, lin28, jub, entpd4, dus11, ddx19b, arid3b, arid3a, apln, abtb1 | up | na | na | [24] |
| miR-125b | up | up | na | |||
| miR-126 | miR-126 | vam1 | up | down | na | [24] |
| miR-130a | up | down | na | |||
| miR-130b | up | down | na | |||
| miR-133a | miR-133 | srf, ptp2, kcne1, hcn2, hcn4, erg, casp9, nfatc4 | up | up | na | [24,34,35,36] |
| miR-133b | up | up | na | |||
| miR-140 | up | na | na | |||
| miR-143 | miR-143 | mapk7, mapk12, aduccine3 | up | up | na | [24,37] |
| miR-145 | miR-145 | irs-1, flj21308, dab2 | up | up | na | [24,38] |
| miR-181a | up | up | na | |||
| miR-181b | up | up | na | |||
| miR-183 | up | up | na | |||
| miR-190 | up | up | na | |||
| miR-191 | up | up | na | |||
| miR-198 * | up | up | na | |||
| miR-202 | up | up | na | |||
| miR-210 | miR-210 | efna3 | up | up | na | [24] |
| miR-298 | up | na | equal | |||
| miR-320 | up | na | na | |||
| miR-322 ** | miR-322 | na | up | na | up | |
| miR-324 | up | down | na | |||
| miR-351 ** | up | na | equal | |||
| miR-373 * | up | down | na | |||
| miR-422b | up | na | na | |||
| miR-453 | up | up | na | |||
| miR-455 | up | na | na | |||
| miR-494 | miR-494 | na | up | up | down | |
| miR-494 | up | na | na | |||
| miR-503 | up | na | down | |||
| miR-513 ** | up | na | na | |||
| miR-517 ** | up | na | na | |||
| miR-518c | up | na | na | |||
| miR-546 * | up | na | equal |
4. A Meta-Analysis of MicroRNA Microarrays in Cardiogenesis and Cardiac Aging
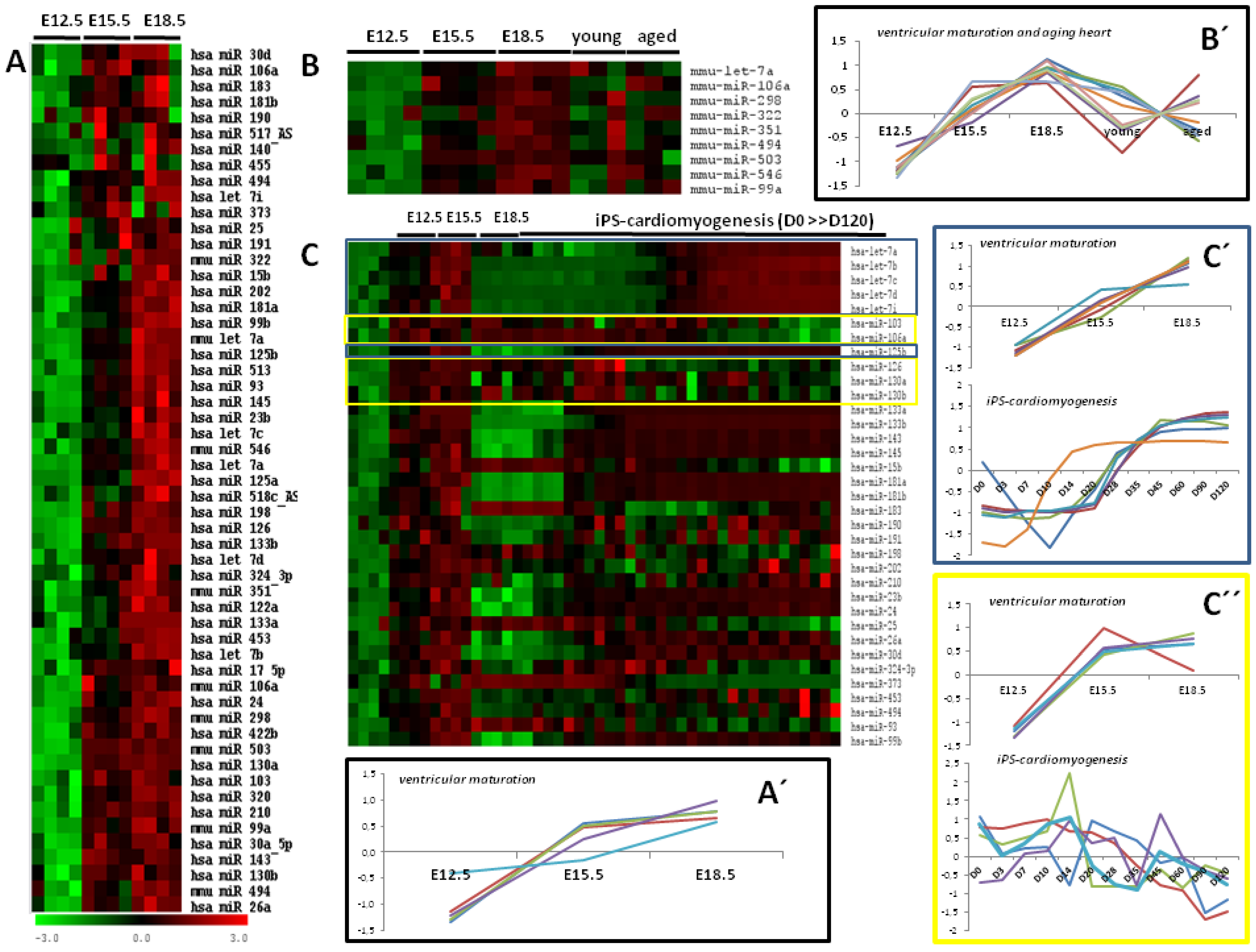
5. The Cardiovascular Role of Differentially Expressed MicroRNAs during Ventricular Development
6. Conclusions & Perspectives
Acknowledgments
Conflict of interest
References
- Kelly, R.G. The second heart field. Curr. Top. Dev. Biol. 2012, 100, 33–65. [Google Scholar] [CrossRef]
- Prall, O.W.; Menon, M.K.; Solloway, M.J.; Watanabe, Y; Zaffran, S.; Bajolle, F.; Biben, C.; McBride, J.J.; Robertson, B.R.; Chaulet, H.; et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 2007, 128, 947–959. [Google Scholar]
- De Castro, M.P.; Acosta, L.; Domínguez, J.N.; Aránega, A.; Franco, D. Molecular diversity of the developing and adult myocardium: Implications for tissue targeting. Curr. Drug. Targets Cardiovasc. Haematol. Disord. 2003, 3, 227–239. [Google Scholar] [CrossRef]
- Chinchilla, A.; Franco, D. Regulatory mechanisms of cardiac development and repair. Cardiovasc. Hematol. Disord. Drug. Targets 2006, 6, 101–112. [Google Scholar]
- Kruithof, B.P.; Duim, S.N.; Moerkamp, A.T.; Goumans, M.J. TGFβ and BMP signaling in cardiac cushion formation: Lessons from mice and chicken. Differentiation 2012, 84, 89–102. [Google Scholar] [CrossRef]
- De Vlaming, A.; Sauls, K.; Hajdu, Z.; Visconti, R.P.; Mehesz, A.N.; Levine, R.A.; Slaugenhaupt, S.A.; Hagège, A.; Chester, A.H.; Markwald, R.R.; et al. Atrioventricular valve development: New perspectives on an old theme. Differentiation 2012, 84, 103–116. [Google Scholar] [CrossRef]
- Männer, J.; Pérez-Pomares, J.M.; Macías, D.; Muñoz-Chápuli, R. The origin, formation and developmental significance of the epicardium: A review. Cells Tissues Organs 2001, 169, 89–103. [Google Scholar] [CrossRef]
- Wessels, A.; Pérez-Pomares, J.M. The epicardium and epicardially derived cells (EPDCs) as cardiac stem cells. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2004, 276, 43–57. [Google Scholar] [CrossRef]
- Liu, N.; Olson, E.N. MicroRNA regulatory networks in cardiovascular development. Dev. Cell 2010, 18, 510–525. [Google Scholar] [CrossRef]
- Bauersachs, J.; Thum, T. Biogenesis and regulation of cardiovascular microRNAs. Circ. Res. 2011, 109, 334–347. [Google Scholar] [CrossRef]
- Gene Expression Omnibus. Available online: http://www.ncbi.nlm.nih.gov/geo/ (accessed on 16 February 2013).
- Zhang, X.; Azhar, G.; Wei, J.Y. The expression of microRNA and microRNA clusters in the aging heart. PLoS One 2012, 7, e34688. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Ravon, M.; Sridhar, S.; Ravindran, P.; Swanson, B.; Bitter, H.; Weiser, T.; Chiao, E.; Certa, U.; Kolaja, K.L. Determination of the human cardiomyocyte mRNA and miRNA differentiation network by fine-scale profiling. Stem Cells Dev. 2012, 21, 1956–1965. [Google Scholar] [CrossRef]
- Chinchilla, A.; Lozano, E.; Daimi, H.; Esteban, F.J.; Crist, C.; Aranega, A.E.; Franco, D. MicroRNA profiling during mouse ventricular maturation: A role for miR-27 modulating Mef2c expression. Cardiovasc. Res. 2011, 89, 98–108. [Google Scholar]
- Bioconductor Limma Package. Available online: http://www.bioconductor.org (accessed on 16 February 2013).
- R Software. Available online: http://www.r-project.org (accessed on 16 February 2013).
- TM4 Software Suite. Available online: http://www.tm4.org (accessed on 16 February 2013).
- Porrello, E.R.; Mahmoud, A.I.; Simpson, E.; Johnson, B.A.; Grinsfelder, D.; Canseco, D.; Mammen, P.P.; Rothermel, B.A.; Olson, E.N.; Sadek, H.A. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. USA 2013, 110, 187–192. [Google Scholar]
- Zhao, Y.; Ransom, J.F.; Li, A.; Vedantham, V.; von Drehle, M.; Muth, A.N.; Tsuchihashi, T.; McManus, M.T.; Schwartz, R.J.; Srivastava, D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 2007, 129, 303–317. [Google Scholar] [CrossRef]
- Saxena, A.; Tabin, C.J. miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proc. Natl. Acad. Sci. USA 2010, 107, 87–91. [Google Scholar] [CrossRef]
- Fish, J.E.; Santoro, M.M.; Morton, S.U.; Yu, S.; Yeh, R.F.; Wythe, J.D.; Ivey, K.N.; Bruneau, B.G.; Stainier, D.Y.; Srivastava, D. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell 2008, 15, 272–284. [Google Scholar] [CrossRef]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar]
- Liu, N.; Bezprozvannaya, S.; Williams, A.H.; Qi, X.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008, 22, 3242–3254. [Google Scholar] [CrossRef]
- Papadopoulos, G.L.; Reczko, M.; Simossis, V.A.; Sethupathy, P.; Hatzigeorgiou, A.G. The database of experimentally supported targets: A functional update of TarBase. Nucl. Acids. Res. 2009, 37, D155–D158. [Google Scholar] [CrossRef]
- Xiang, R.; Lei, H.; Chen, M.; Li, Q.; Sun, H.; Ai, J.; Chen, T.; Wang, H.; Fang, Y.; Zhou, Q. The miR-17-92 cluster regulates FOG-2 expression and inhibits proliferation of mouse embryonic cardiomyocytes. Braz. J. Med. Biol. Res. 2012, 45, 131–138. [Google Scholar] [CrossRef]
- Sirish, P.; López, J.E.; Li, N.; Wong, A.; Timofeyev, V.; Young, J.N.; Majdi, M.; Li, R.A.; Chen, H.S.; Chiamvimonvat, N. MicroRNA profiling predicts a variance in the proliferative potential of cardiac progenitor cells derived from neonatal and adult murine hearts. J. Mol. Cell Cardiol. 2012, 52, 264–272. [Google Scholar] [CrossRef]
- Lagendijk, A.K.; Goumans, M.J.; Burkhard, S.B.; Bakkers, J. MicroRNA-23 restricts cardiac valve formation by inhibiting Has2 and extracellular hyaluronic acid production. Circ. Res. 2011, 109, 649–657. [Google Scholar] [CrossRef]
- Fiedler, J.; Jazbutyte, V.; Kirchmaier, B.C.; Gupta, S.K.; Lorenzen, J.; Hartmann, D.; Galuppo, P.; Kneitz, S.; Pena, J.T.; Sohn-Lee, C.; et al. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation 2011, 124, 720–730. [Google Scholar] [CrossRef]
- Li, D.F.; Tian, J.; Guo, X.; Huang, L.M.; Xu, Y.; Wang, C.C.; Wang, J.F.; Ren, A.J.; Yuan, W.J.; Lin, L. Induction of microRNA-24 by HIF-1 protects against ischemic injury in rat cardiomyocytes. Physiol. Res. 2013, 61, 555–565. [Google Scholar]
- Vacchi-Suzzi, C.; Hahne, F.; Scheubel, P.; Marcellin, M.; Dubost, V.; Westphal, M.; Boeglen, C.; Büchmann-Møller, S.; Cheung, M.S.; Cordier, A.; et al. Heart structure-specific transcriptomic atlas reveals conserved microRNA-mRNA interactions. PLoS One 2013, 8, e52442. [Google Scholar] [CrossRef]
- Duisters, R.F.; Tijsen, A.J.; Schroen, B.; Leenders, J.J.; Lentink, V.; van der Made, I.; Herias, V.; van Leeuwen, R.E.; Schellings, M.W.; Barenbrug, P.; et al. miR-133 and miR-30 regulate connective tissue growth factor: Implications for a role of microRNAs in myocardial matrix remodeling. Circ. Res. 2009, 104, 170–178. [Google Scholar] [CrossRef]
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar]
- Huang, X.; Huang, F.; Yang, D.; Dong, F.; Shi, X.; Wang, H.; Zhou, X.; Wang, S.; Dai, S. Expression of microRNA-122 contributes to apoptosis in H9C2 myocytes. J. Cell. Mol. Med. 2012, 16, 2637–2646. [Google Scholar] [CrossRef]
- Li, Q.; Lin, X.; Yang, X.; Chang, J. NFATc4 is negatively regulated in miR-133a-mediated cardiomyocyte hypertrophic repression. Am. J. Physiol. Heart Circ. Physiol. 2010, 298, H1340–H1347. [Google Scholar] [CrossRef]
- Dong, D.L.; Chen, C.; Huo, R.; Wang, N.; Li, Z.; Tu, Y.J.; Hu, J.T.; Chu, X.; Huang, W.; Yang, B.F. Reciprocal repression between microRNA-133 and calcineurin regulates cardiac hypertrophy: A novel mechanism for progressive cardiac hypertrophy. Hypertension 2010, 55, 946–952. [Google Scholar] [CrossRef]
- Horie, T.; Ono, K.; Nishi, H.; Iwanaga, Y.; Nagao, K.; Kinoshita, M.; Kuwabara, Y.; Takanabe, R.; Hasegawa, K.; Kita, T.; et al. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochem. Biophys. Res. Commun. 2009, 389, 315–320. [Google Scholar] [CrossRef]
- Deacon, D.C.; Nevis, K.R.; Cashman, T.J.; Zhou, Y.; Zhao, L.; Washko, D.; Guner-Ataman, B.; Burns, C.G.; Burns, C.E. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis. Development 2010, 137, 1887–1896. [Google Scholar] [CrossRef]
- Mayorga, M.E.; Penn, M.S. miR-145 is differentially regulated by TGF-β1 and ischaemia and targets Disabled-2 expression and wnt/β-catenin activity. J. Cell. Mol. Med. 2012, 16, 1106–1113. [Google Scholar] [CrossRef]
- Cao, L.; Kong, L.P.; Yu, Z.B.; Han, S.P.; Bai, Y.F.; Zhu, J.; Hu, X.; Zhu, C.; Zhu, S.; Guo, X.R. MicroRNA expression profiling of the developing mouse heart. Int. J. Mol. Med. 2012, 30, 1095–1104. [Google Scholar]
- Han, M.; Toli, J.; Abdellatif, M. MicroRNAs in the cardiovascular system. Curr. Opin. Cardiol. 2011, 26, 181–189. [Google Scholar] [CrossRef]
- Malizia, A.P.; Wang, D.Z. MicroRNAs in cardiomyocyte development. WIREs Syst. Biol. Med. 2011, 3, 183–190. [Google Scholar]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef]
- Bostjancic, E.; Zidar, N.; Stajer, D.; Glavac, D. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 are dysregulated in human myocardial infarction. Cardiology 2010, 115, 163–169. [Google Scholar] [CrossRef]
- Bostjancic, E.; Zidar, N.; Stajer, D.; Glavac, D. MicroRNA miR-1 is up-regulated in remote myocardium in patients with myocardial infarction. Folia Biol. (Praha) 2010, 56, 27–31. [Google Scholar]
- Belevych, A.E.; Sansom, S.E.; Terentyeva, R.; Ho, H.T.; Nishijima, Y.; Martin, M.M.; Jindal, H.K.; Rochira, J.A.; Kunitomo, Y.; Abdellatif, M.; et al. MicroRNA-1 and -133 increase arrhythmogenesis in heart failure by dissociating phosphatase activity from RyR2 complex. PLoS One 2011, 6, e28324. [Google Scholar] [CrossRef]
- Li, H.; Li, S.; Yu, B.; Liu, S. Expression of miR-133 and miR-30 in chronic atrial fibrillation in canines. Mol. Med. Rep. 2012, 5, 1457–1460. [Google Scholar]
- Villar, A.V.; Merino, D.; Wenner, M.; Llano, M.; Cobo, M.; Montalvo, C.; García, R.; Martín-Durán, R.; Hurlé, J.M.; Hurlé, M.A.; et al. Myocardial gene expression of microRNA-133a and myosin heavy and light chains, in conjunction with clinical parameters, predict regression of left ventricular hypertrophy after valve replacement in patients with aortic stenosis. Heart 2011, 97, 1132–1137. [Google Scholar]
- Yin, V.P.; Lepilina, A.; Smith, A.; Poss, K.D. Regulation of zebrafish heart regeneration by miR-133. Dev. Biol. 2012, 365, 319–327. [Google Scholar] [CrossRef]
- Miyasaka, K.Y.; Kida, Y.S.; Banjo, T.; Ueki, Y.; Nagayama, K.; Matsumoto, T.; Sato, M.; Ogura, T. Heartbeat regulates cardiogenesis by suppressing retinoic acid signaling via expression of miR-143. Mech. Dev. 2011, 128, 18–28. [Google Scholar] [CrossRef]
- Hsu, J.; Hanna, P.; van Wagoner, D.R.; Barnard, J.; Serre, D.; Chung, M.K.; Smith, J.D. Whole genome expression differences in human left and right atria ascertained by RNA sequencing. Circ. Cardiovasc. Genet. 2012, 5, 327–335. [Google Scholar] [CrossRef]
- Caruso, P.; MacLean, M.R.; Khanin, R.; McClure, J.; Soon, E.; Southgate, M.; MacDonald, R.A.; Greig, J.A.; Robertson, K.E.; Masson, R.; et al. Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 716–723. [Google Scholar] [CrossRef]
- Meder, B.; Keller, A.; Vogel, B.; Haas, J.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Just, S.; Borries, A.; Rudloff, J.; Leidinger, P.; et al. MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res. Cardiol. 2011, 106, 13–23. [Google Scholar] [CrossRef]
- Fichtlscherer, S.; De Rosa, S.; Fox, H.; Schwietz, T.; Fischer, A.; Liebetrau, C.; Weber, M.; Hamm, C.W.; Röxe, T.; Müller-Ardogan, M.; et al. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 2010, 107, 677–684. [Google Scholar] [CrossRef]
- Li, R.; Yan, G.; Li, Q.; Sun, H.; Hu, Y.; Sun, J.; Xu, B. MicroRNA-145 protects cardiomyocytes against hydrogen peroxide (H2O2)-induced apoptosis through targeting the mitochondria apoptotic pathway. PLoS One 2012, 7, e44907. [Google Scholar] [CrossRef]
- Ventura, A.; Young, A.G.; Winslow, M.M.; Lintault, L.; Meissner, A.; Erkeland, S.J.; Newman, J.; Bronson, R.T.; Crowley, D.; Stone, J.R.; et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008, 132, 875–886. [Google Scholar]
- Danielson, L.S.; Park, D.S.; Rotllan, N.; Chamorro-Jorganes, A.; Guijarro, M.V.; Fernandez-Hernando, C.; Fishman, G.I.; Phoon, C.K.; Hernando, E. Cardiovascular dysregulation of miR-17–92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J. 2012. [Google Scholar] [CrossRef]
- Van Almen, G.C.; Verhesen, W.; van Leeuwen, R.E.; van de Vrie, M.; Eurlings, C.; Schellings, M.W.; Swinnen, M.; Cleutjens, J.P.; van Zandvoort, M.A.; Heymans, S.; et al. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell 2011, 10, 769–779. [Google Scholar] [CrossRef]
- Lee, C.; Mitsialis, S.A.; Aslam, M.; Vitali, S.H.; Vergadi, E.; Konstantinou, G.; Sdrimas, K.; Fernandez-Gonzalez, A.; Kourembanas, S. Exosomes mediate the cytoprotective action of mesenchymal stromal cells on hypoxia-induced pulmonary hypertension. Circulation 2012, 126, 2601–2611. [Google Scholar] [CrossRef]
- Pullamsetti, S.S.; Doebele, C.; Fischer, A.; Savai, R.; Kojonazarov, B.; Dahal, B.K.; Ghofrani, H.A.; Weissmann, N.; Grimminger, F.; Bonauer, A.; et al. Inhibition of microRNA-17 improves lung and heart function in experimental pulmonary hypertension. Am. J. Respir. Crit. Care Med. 2012, 185, 409–419. [Google Scholar]
- Shan, S.W.; Lee, D.Y.; Deng, Z.; Shatseva, T.; Jeyapalan, Z.; Du, W.W.; Zhang, Y.; Xuan, J.W.; Yee, S.P.; Siragam, V.; et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat. Cell Biol. 2009, 11, 1031–1038. [Google Scholar]
- Han, M.; Toli, J.; Abdellatif, M. MicroRNAs in the cardiovascular system. Curr. Opin. Cardiol. 2011, 26, 181–189. [Google Scholar] [CrossRef]
- Mutharasan, R.K.; Nagpal, V.; Ichikawa, Y.; Ardehali, H. microRNA-210 is upregulated in hypoxic cardiomyocytes through Akt- and p53-dependent pathways and exerts cytoprotective effects. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H1519–H1530. [Google Scholar] [CrossRef]
- Porrello, E.R.; Johnson, B.A.; Aurora, A.B.; Simpson, E.; Nam, Y.J.; Matkovich, S.J.; Dorn, G.W.; van Rooij, E.; Olson, E.N. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 2011, 109, 670–679. [Google Scholar] [CrossRef]
- Divakaran, V.; Adrogue, J.; Ishiyama, M.; Entman, M.L.; Haudek, S.; Sivasubramanian, N.; Mann, D.L. Adaptive and maladptive effects of SMAD3 signaling in the adult heart after hemodynamic pressure overloading. Circ. Heart Fail. 2009, 2, 633–642. [Google Scholar] [CrossRef]
- Wang, J.; Huang, W.; Xu, R.; Nie, Y.; Cao, X.; Meng, J.; Xu, X.; Hu, S.; Zheng, Z. MicroRNA-24 regulates cardiac fibrosis after myocardial infarction. J. Cell Mol. Med. 2012, 16, 2150–2160. [Google Scholar] [CrossRef]
- Hu, S.; Huang, M.; Nguyen, P.K.; Gong, Y.; Li, Z.; Jia, F.; Lan, F.; Liu, J.; Nag, D.; Robbins, R.C.; Wu, J.C. Novel microRNA prosurvival cocktail for improving engraftment and function of cardiac progenitor cell transplantation. Circulation 2011, 124, S27–S34. [Google Scholar] [CrossRef]
- Fish, J.E.; Srivastava, D. MicroRNAs: Opening a new vein in angiogenesis research. Sci. Signal 2009, 2. [Google Scholar] [CrossRef]
- Bang, C.; Fiedler, J.; Thum, T. Cardiovascular importance of the microRNA-23/27/24 family. Microcirculation 2012, 19, 208–214. [Google Scholar] [CrossRef]
- Stankunas, K.; Ma, G.K.; Kuhnert, F.J.; Kuo, C.J.; Chang, C.P. VEGF signaling has distinct spatiotemporal roles during heart valve development. Dev. Biol. 2010, 347, 325–336. [Google Scholar] [CrossRef]
- Shi, H.; Chen, L.; Wang, H.; Zhu, S.; Dong, C.; Webster, K.A.; Wei, J. Synergistic induction of miR-126 by hypoxia and HDAC inhibitors in cardiac myocytes. Biochem. Biophys. Res. Commun. 2013, 430, 827–832. [Google Scholar] [CrossRef]
- Tzur, G.; Levy, A.; Meiri, E.; Barad, O.; Spector, Y.; Bentwich, Z.; Mizrahi, L.; Katzenellenbogen, M.; Ben-Shushan, E.; Reubinoff, B.E.; Galun, E. MicroRNA expression patterns and function in endodermal differentiation of human embryonic stem cells. PLoS One 2008, 3, e3726. [Google Scholar] [CrossRef]
- Stuckenholz, C.; Lu, L.; Thakur, P.; Kaminski, N.; Bahary, N. FACS-assisted microarray profiling implicates novel genes and pathways in zebrafish gastrointestinal tract development. Gastroenterology 2009, 137, 1321–1332. [Google Scholar] [CrossRef]
- Xu, H.; He, J.H.; Xiao, Z.D.; Zhang, Q.Q.; Chen, Y.Q.; Zhou, H.; Qu, L.H. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology 2010, 52, 1431–1442. [Google Scholar] [CrossRef]
- Laudadio, I.; Manfroid, I.; Achouri, Y.; Schmidt, D.; Wilson, M.D.; Cordi, S.; Thorrez, L.; Knoops, L.; Jacquemin, P.; Schuit, F.; et al. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology 2012, 142, 119–129. [Google Scholar]
- Wong, S.S.; Ritner, C.; Ramachandran, S.; Aurigui, J.; Pitt, C.; Chandra, P.; Ling, V.B.; Yabut, O.; Bernstein, H.S. miR-125b promotes early germ layer specification through Lin28/let-7d and preferential differentiation of mesoderm in human embryonic stem cells. PLoS One 2012, 7, e36121. [Google Scholar] [CrossRef]
- Ahmed, R.P.; Haider, H.K.; Buccini, S.; Li, L.; Jiang, S.; Ashraf, M. Reprogramming of skeletal myoblasts for induction of pluripotency for tumor-free cardiomyogenesis in the infarcted heart. Circ. Res. 2011, 109, 60–70. [Google Scholar] [CrossRef]
- Fu, J.; Peng, C.; Wang, W.; Jin, H.; Tang, Q.; Wei, X. Let-7g is involved in doxorubicin induced myocardial injury. Environ. Toxicol. Pharmacol. 2012, 33, 312–317. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Bonet, F.; Hernandez-Torres, F.; Esteban, F.J.; Aranega, A.; Franco, D. Comparative Analyses of MicroRNA Microarrays during Cardiogenesis: Functional Perspectives. Microarrays 2013, 2, 81-96. https://doi.org/10.3390/microarrays2020081
Bonet F, Hernandez-Torres F, Esteban FJ, Aranega A, Franco D. Comparative Analyses of MicroRNA Microarrays during Cardiogenesis: Functional Perspectives. Microarrays. 2013; 2(2):81-96. https://doi.org/10.3390/microarrays2020081
Chicago/Turabian StyleBonet, Fernando, Francisco Hernandez-Torres, Franciso J. Esteban, Amelia Aranega, and Diego Franco. 2013. "Comparative Analyses of MicroRNA Microarrays during Cardiogenesis: Functional Perspectives" Microarrays 2, no. 2: 81-96. https://doi.org/10.3390/microarrays2020081
APA StyleBonet, F., Hernandez-Torres, F., Esteban, F. J., Aranega, A., & Franco, D. (2013). Comparative Analyses of MicroRNA Microarrays during Cardiogenesis: Functional Perspectives. Microarrays, 2(2), 81-96. https://doi.org/10.3390/microarrays2020081







