Development and Optimization of a Thrombin Sandwich Aptamer Microarray
Abstract
:1. Introduction
2. Materials and Methods
2.1. Aptamers and Proteins Solutions
2.2. Aptamer Preparation for Microarray Printing
2.3. Aptamer Microarray Printing
2.4. Detection Layer Preparation
2.5. Sandwich Aptamer Microarray Assays
2.6. Slides Scanning and Data Analysis
3. Results and Discussion
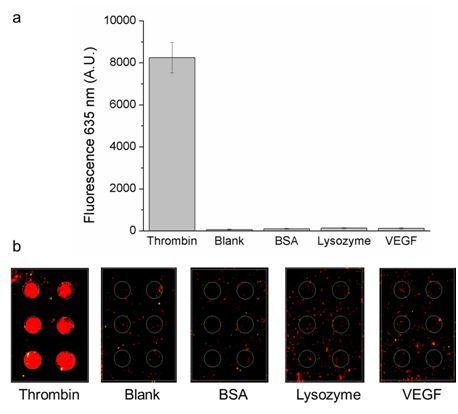
3.1. Thrombin Specificity
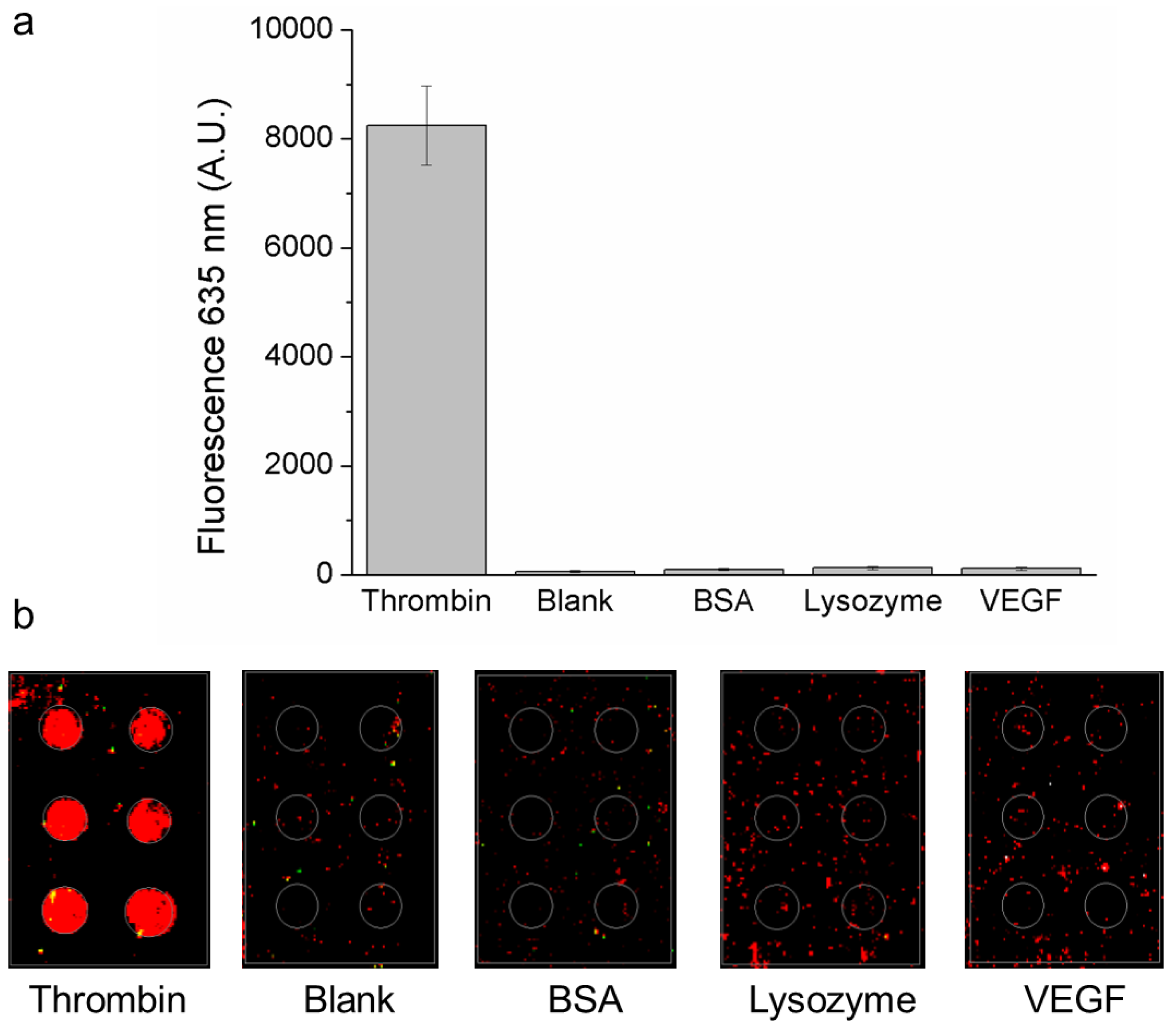
3.2. Effect of Incubation Time
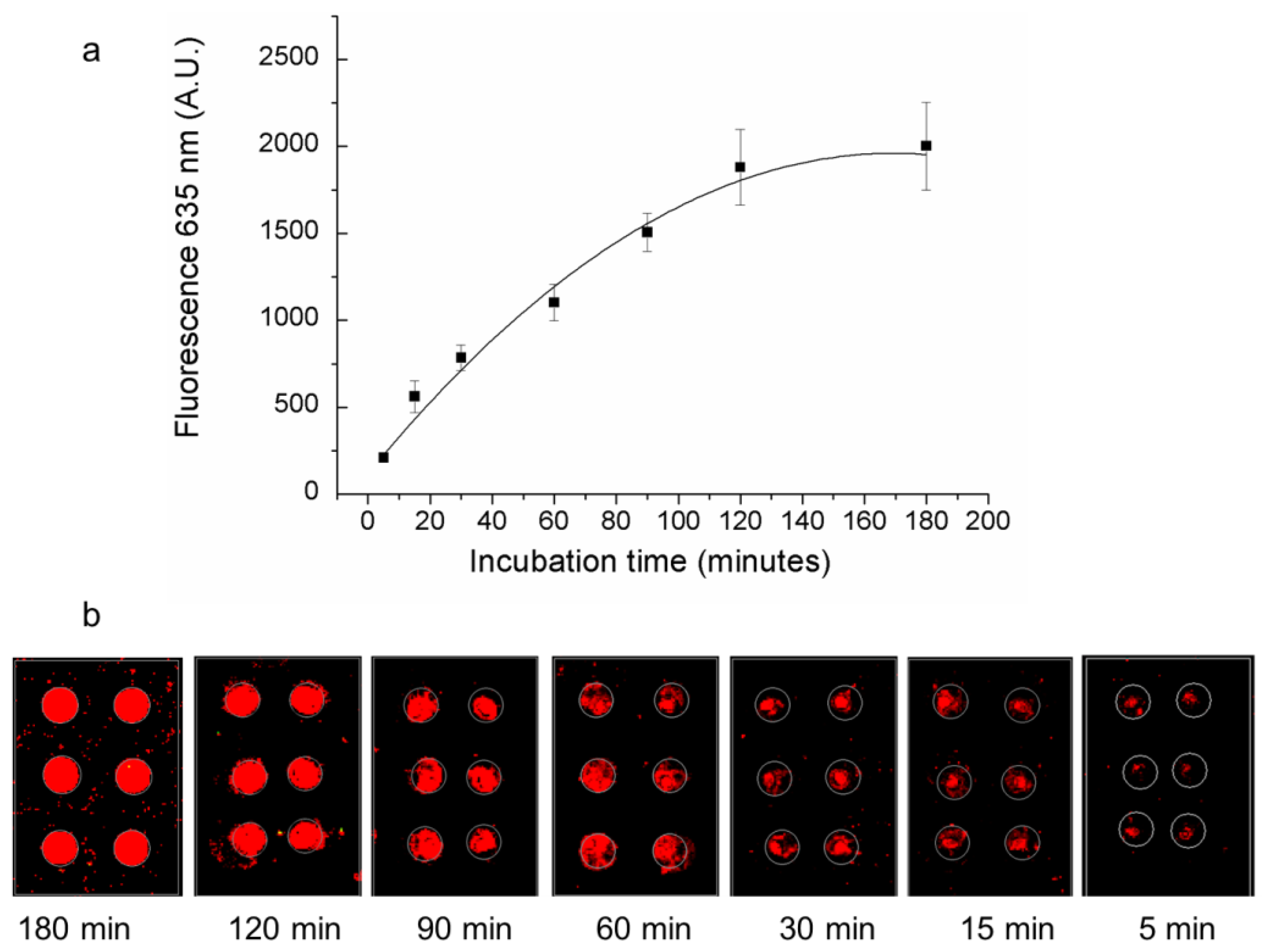
3.3. Effect of Serum
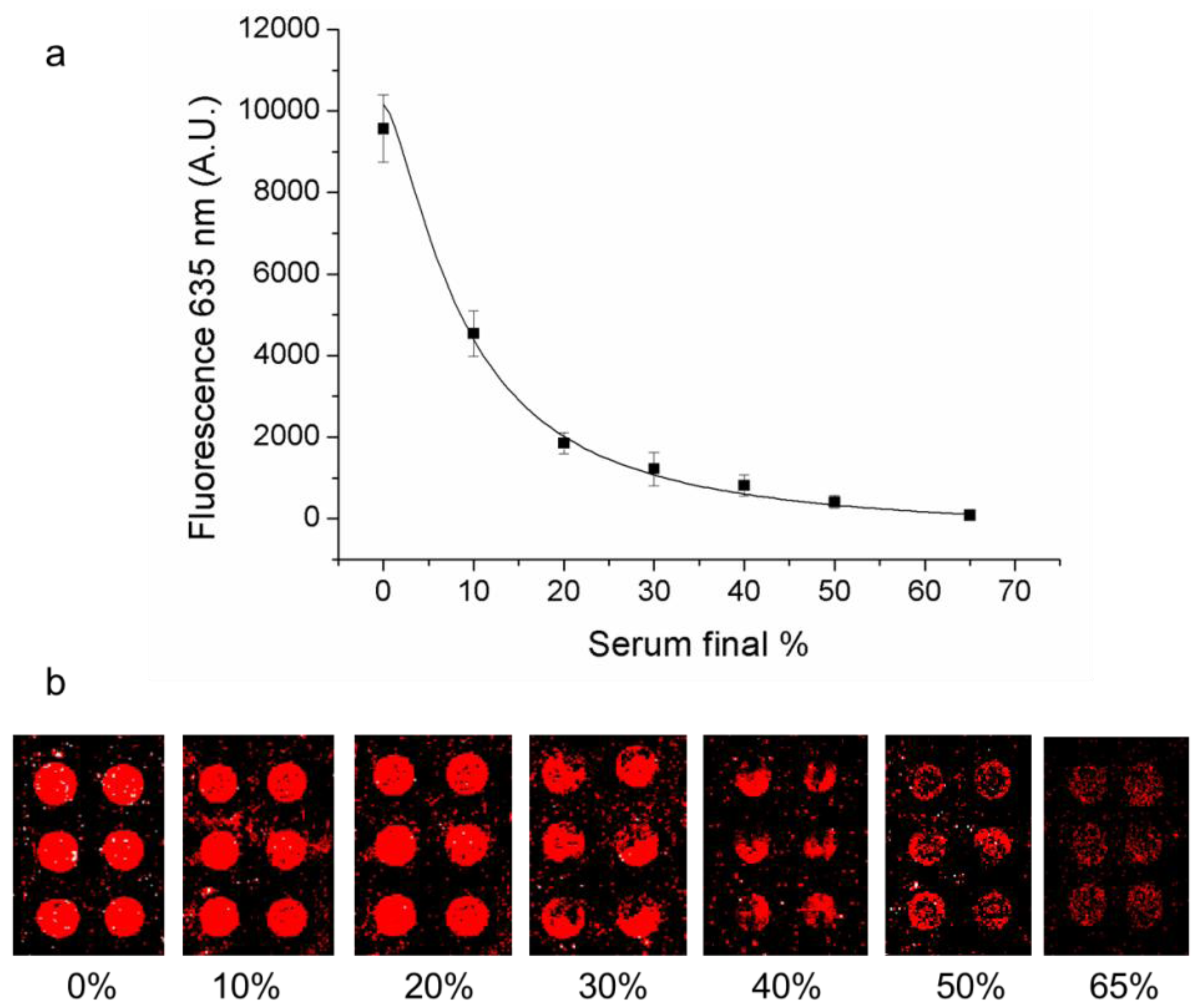
3.4. Limit of Detection of Direct and Indirect Methods
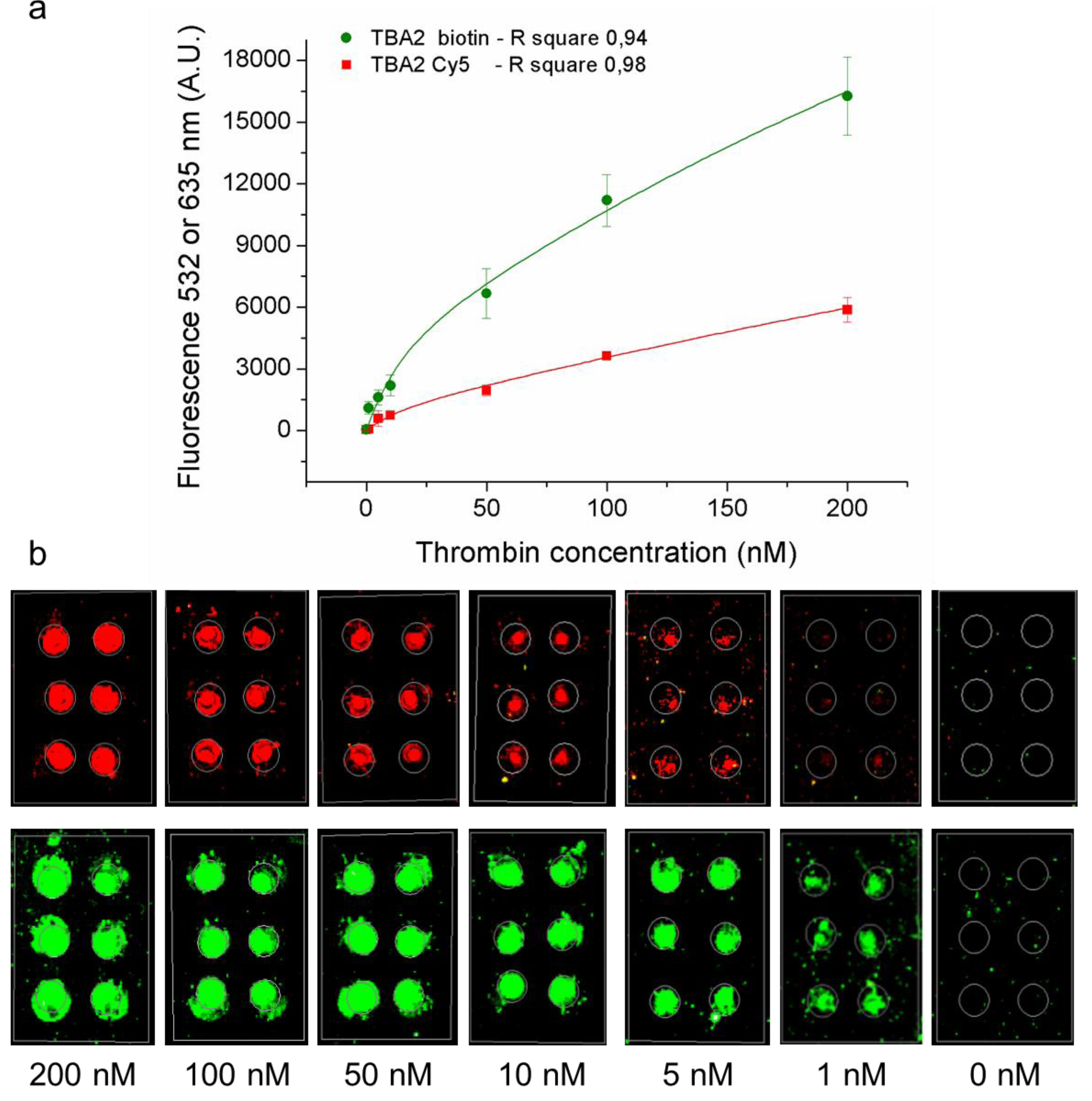
3.5. Indirect Method for Aptasensor Detection: Effect of Serum

4. Conclusions
Acknowledgments
References
- Fenton, J.W., II. Thrombin specificity. Ann. N. Y. Acad Sci. 1981, 370, 468–495. [Google Scholar] [CrossRef]
- Shuman, M.A. Thrombin-cellular interactions. Ann. N. Y. Acad Sci. 1986, 485, 228–239. [Google Scholar] [CrossRef]
- Shuman, M.A.; Majerus, P.W. The measurement of thrombin in clotting blood by radioimmunoassay. J. Clin. Invest. 1976, 58, 1249–1258. [Google Scholar]
- Becker, R.C.; Spencer, F.A. Thrombin: Structure, biochemistry, measurement, and status in clinical medicine. J. Thrombosis Thrombolysis 1998, 5, 215–229. [Google Scholar] [CrossRef]
- Suo, Z.; Citron, B.A.; Festoff, B.W. Thrombin: A potential proinflammatory mediator in neurotrauma and neurodegenerative disorders. Curr. Drug Targets Inflamm. Allergy 2004, 3, 105–114. [Google Scholar] [CrossRef]
- Turgeon, V.L.; Houenou, L.J. The role of thrombin-like (serine) proteases in the development, plasticity and pathology of the nervous system. Brain Res. Rev. 1997, 25, 85–95. [Google Scholar]
- Cho, E.J.; Lee, J.W.; Ellington, A.D. Applications of aptamers as sensors. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2009, 2, 241–264. [Google Scholar]
- Khati, M. The future of aptamers in medicine. J. Clin. Pathol. (Lond.) 2010, 63, 480–487. [Google Scholar]
- Famulok, M.; Mayer, G. Aptamer modules as sensors and detectors. Acc. Chem. Res. 2011, 44, 1349–1358. [Google Scholar] [CrossRef]
- Paborsky, L.R.; McCurdy, S.N.; Griffin, L.C.; Toole, J.J.; Leung, L.L. The single-stranded DNA aptamer-binding site of human thrombin. J. Biol. Chem. 1993, 268, 20808–20811. [Google Scholar]
- Tasset, D.M.; Kubik, M.F.; Steiner, W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 1997, 272, 688–698. [Google Scholar] [CrossRef]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745–3749. [Google Scholar] [CrossRef]
- Ponikova, S.; Antalik, M.; Hianik, T. A circular dichroism study of the stability of guanine quadruplexes of thrombin DNA aptamers at presence of K+ and Na+ ions. Gen. Physiol. Biophys. 2008, 27, 271–277. [Google Scholar]
- Ikebukuro, K.; Kiyohara, C.; Sode, K. Novel electrochemical sensor system for protein using the aptamers in sandwich manner. Biosens. Bioelectron. 2005, 20, 2168–2172. [Google Scholar] [CrossRef]
- Centi, S.; Tombelli, S.; Minunni, M.; Mascini, M. Aptamer-based detection of plasma proteins by an electrochemical assay coupled to magnetic beads. Anal. Chem. 2007, 79, 1466–1473. [Google Scholar]
- Tang, Q.; Su, X.; Loh, K.P. Surface plasmon resonance spectroscopy study of interfacial binding of thrombin to antithrombin DNA aptamers. J. Colloid Interface Sci. 2007, 315, 99–106. [Google Scholar] [CrossRef]
- Fang, L.; Lü, Z.; Wei, H.; Wang, E. A electrochemiluminescence aptasensor for detection of thrombin incorporating the capture aptamer labeled with gold nanoparticles immobilized onto the thio-silanized ITO electrode. Anal. Chim. Acta 2008, 628, 80–86. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Y.; Liu, C.; Huang, J.; Yang, P.; Liu, B. Microfluidic chip-based aptasensor for amplified electrochemical detection of human thrombin. Electrochem. Comm. 2010, 12, 258–261. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, X.-F.; Shao, Y.; Le, X.C. Aptamer-based affinity chromatographic assays for thrombin. Anal. Chem. 2008, 80, 7586–7593. [Google Scholar]
- Edwards, K.A.; Wang, Y.; Baeumner, A.J. Aptamer sandwich assays: Human α-thrombin detection using liposome enhancement. Anal. Bioanal. Chem. 2010, 398, 2645–2654. [Google Scholar]
- Chen, J.; Zhang, J.; Li, J.; Yang, H.-H.; Fu, F.; Chen, G. An ultrasensitive signal-on electrochemical aptasensor via target-induced conjunction of split aptamer fragments. Biosens. Bioelectron. 2010, 25, 996–1000. [Google Scholar] [CrossRef]
- Tennico, Y.H.; Hutanu, D.; Koesdjojo, M.T.; Bartel, C.M.; Remcho, V.T. On-chip aptamer-based sandwich assay for thrombin detection employing magnetic beads and quantum dots. Anal. Chem. 2010, 82, 5591–5597. [Google Scholar]
- Lao, Y.-H.; Peck, K.; Chen, L.-C. Enhancement of aptamer microarray sensitivity through spacer optimization and avidity effect. Anal. Chem. 2009, 81, 1747–1754. [Google Scholar] [CrossRef]
- Zhao, Q.; Lu, X.; Yuan, C.-G.; Li, X.-F.; Le, X.C. Aptamer-linked assay for thrombin using gold nanoparticle amplification and inductively coupled plasma-mass spectrometry detection. Anal. Chem. 2009, 81, 7484–7489. [Google Scholar] [CrossRef]
- Niu, S.; Qu, L.; Zhang, Q.; Lin, J. Fluorescence detection of thrombin using autocatalytic strand displacement cycle reaction and a dual-aptamer DNA sandwich assay. Anal. Biochem. 2012, 421, 362–367. [Google Scholar]
- Wang, L.; Li, L.; Xu, Y.; Cheng, G.; He, P.; Fang, Y. Simultaneously fluorescence detecting thrombin and lysozyme based on magnetic nanoparticle condensation. Talanta 2009, 79, 557–561. [Google Scholar] [CrossRef]
- Sosic, A.; Meneghello, A.; Cretaio, E.; Gatto, B. Human thrombin detection through a sandwich aptamer microarray: Interaction analysis in solution and in solid phase. Sensors 2011, 11, 9426–9441. [Google Scholar] [CrossRef]
- Yang, C.; Wang, Y.; Marty, J.-L.; Yang, X. Aptamer-based colorimetric biosensing of Ochratoxin A using unmodified gold nanoparticles indicator. Biosens. Bioelectron. 2011, 26, 2724–2727. [Google Scholar]
- Sahni, A.; Francis, C.W. Vascular endothelial growth factor binds to fibrinogen and fibrin and stimulates endothelial cell proliferation. Blood 2000, 96, 3772–3778. [Google Scholar]
- Keyt, B.A.; Berleau, L.T.; Nguyen, H.V.; Chen, H.; Heinsohn, H.; Vandlen, R.; Ferrara, N. The carboxyl-terminal domain (111–165) of vascular endothelial growth factor is critical for its mitogenic potency. J. Biol. Chem. 1996, 271, 7788–7795. [Google Scholar]
- Hornbeck, P.; Winston, S.E.; Fuller, S.A. Enzyme-linked immunosorbent assays (ELISA). In Current Protocols in Molecular Biology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001. [Google Scholar]
- Lim, Y.C.; Kouzani, A.Z.; Duan, W. Aptasensors: A review. J. Biomed. Nanotechnol. 2010, 6, 93–105. [Google Scholar] [CrossRef]
- Zeng, X.; Shen, Z.; Mernaugh, R. Recombinant antibodies and their use in biosensors. Anal. Bioanal. Chem. 2012, 402, 3027–3038. [Google Scholar]
- Huang, D.W.; Niu, C.G.; Qin, P.Z.; Ruan, M.; Zeng, G.M. Time-resolved fluorescence aptamer-based sandwich assay for thrombin detection. Talanta 2010, 83, 185–189. [Google Scholar] [CrossRef]
- Chen, Y.; Nakamoto, K.; Niwa, O.; Corn, R.M. On-chip synthesis of RNA aptamer microarrays for multiplexed protein biosensing with SPR imaging measurements. Langmuir 2012, 28, 8281–8285. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Meneghello, A.; Sosic, A.; Antognoli, A.; Cretaio, E.; Gatto, B. Development and Optimization of a Thrombin Sandwich Aptamer Microarray. Microarrays 2012, 1, 95-106. https://doi.org/10.3390/microarrays1020095
Meneghello A, Sosic A, Antognoli A, Cretaio E, Gatto B. Development and Optimization of a Thrombin Sandwich Aptamer Microarray. Microarrays. 2012; 1(2):95-106. https://doi.org/10.3390/microarrays1020095
Chicago/Turabian StyleMeneghello, Anna, Alice Sosic, Agnese Antognoli, Erica Cretaio, and Barbara Gatto. 2012. "Development and Optimization of a Thrombin Sandwich Aptamer Microarray" Microarrays 1, no. 2: 95-106. https://doi.org/10.3390/microarrays1020095