Improvement in Classification Performance Based on Target Vector Modification for All-Transfer Deep Learning
Abstract
:1. Introduction
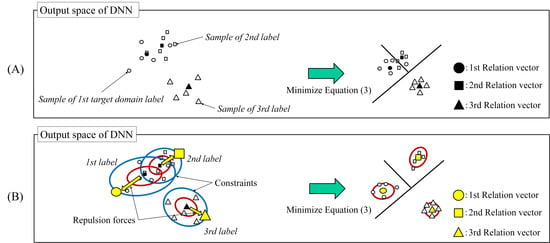
- We propose a relation vector modification for ATDL with constrained pairwise repulsive forces between relation vectors.
- Experimental results showed that our method is effective, especially when the target domain data are significantly small.
- We also showed that the distance between the relation vectors relates to the classification performance.
2. Related Work
2.1. All-Transfer Deep Learning
2.2. Outline
2.2.1. The First DNN Construction (Figure 1A)
2.2.2. Relation Vector Estimation (Figure 1B)
2.2.3. Second DNN Construction (Figure 1C)
3. Target Vector Estimation with a Constrained Pairwise Repulsive Force
3.1. Repulsion Force
3.2. Constraint
3.3. Estimation
4. Experimental Results
4.1. Environment and Parameter Settings
4.2. Simulation Experiments
4.3. 2-DE Images Classification
4.3.1. Comparison with Conventional Methods
4.3.2. Classification Performance of CNN (Convolutional Neural Network)
4.3.3. Comparison of Various Source Tasks
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef]
- Sawada, Y.; Sato, Y.; Nakada, T.; Ujimoto, K.; Hayashi, N. All-Transfer Learning for Deep Neural Networks and Its Application to Sepsis Classification. In Proceedings of the European Conference on Artificial Intelligence, The Hague, The Netherlands, 29 August–2 September 2016; pp. 1586–1587. [Google Scholar]
- Pan, S.J.; Yang, Q. A survey on transfer learning. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Long, M.; Cao, Y.; Wang, J.; Jordan, M. Learning Transferable Features with Deep Adaptation Networks. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 97–105. [Google Scholar]
- Tzeng, E.; Hoffman, J.; Darrell, T.; Saenko, K. Simultaneous deep transfer across domains and tasks. In Proceedings of the International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 4068–4076. [Google Scholar]
- Agrawal, P.; Girshick, R.; Malik, J. Analyzing the performance of multilayer neural networks for object recognition. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 329–344. [Google Scholar]
- Donahue, J.; Jia, Y.; Vinyals, O.; Hoffman, J.; Zhang, N.; Tzeng, E.; Darrell, T. DeCAF: A Deep Convolutional Activation Feature for Generic Visual Recognition. In Proceedings of the International Conference on Machine Learning, Beijing, China, 21–26 June 2014; pp. 647–655. [Google Scholar]
- Oquab, M.; Bottou, L.; Laptev, I.; Sivic, J. Learning and transferring mid-level image representations using convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1717–1724. [Google Scholar]
- Luk, J.M.; Lam, B.Y.; Lee, N.P.; Ho, D.W.; Sham, P.C.; Chen, L.; Peng, J.; Leng, X.; Day, P.J.; Fan, S.T. Artificial neural networks and decision tree model analysis of liver cancer proteomes. Biochem. Biophys. Res. Commun. 2007, 361, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? In Proceedings of the Advances Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; pp. 3320–3328. [Google Scholar]
- Sharif Razavian, A.; Azizpour, H.; Sullivan, J.; Carlsson, S. CNN features off-the-shelf: an astounding baseline for recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Columbus, OH, USA, 23–28 June 2014; pp. 806–813. [Google Scholar]
- Silberman, N.; Guadarrama, S. TensorFlow-Slim Image Classification Model Library. Available online: https://github.com/tensorflow/models/tree/master/research/slim (accessed on 28 November 2018).
- Koitka, S.; Friedrich, C.M. Traditional Feature Engineering and Deep Learning Approaches at Medical Classification Task of ImageCLEF 2016. In Proceedings of the Conference and Labs of the Evaluation Forum, Évora, Portugal, 5–8 September 2016; pp. 304–317. [Google Scholar]
- Mou, L.; Meng, Z.; Yan, R.; Li, G.; Xu, Y.; Zhang, L.; Jin, Z. How Transferable are Neural Networks in NLP Applications? In Proceedings of the Empirical Methods in Natural Language Processing, Austin, TX, USA, 1–5 November 2016; pp. 479–489. [Google Scholar]
- Friedman, J.; Hastie, T.; Tibshirani, R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics 2008, 9, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Gilks, W.R. Markov Chain Monte Carlo; Wiley Online Library: New York, NY, USA, 2005. [Google Scholar]
- Wilson, A.C.; Roelofs, R.; Stern, M.; Srebro, N.; Recht, B. The marginal value of adaptive gradient methods in machine learning. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4148–4158. [Google Scholar]
- Goodfellow, I.J.; Warde-Farley, D.; Lamblin, P.; Dumoulin, V.; Mirza, M.; Pascanu, R.; Bergstra, J.; Bastien, F.; Bengio, Y. Pylearn2: A machine learning research library. arXiv, 2013; arXiv:1308.4214. [Google Scholar]
- Bengio, Y. The consciousness prior. arXiv, 2017; arXiv:1709.08568. [Google Scholar]
- Krizhevsky, A.; Hinton, G. Learning Multiple Layers of Features from Tiny Images. Master’s Thesis, Toronto University, Toronto, ON, Canada, 2009. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the Advances Neural Information Processing Systems, Lake Tahoe, CA and NV, USA, 3–8 December 2012; pp. 1097–1105. [Google Scholar]
- Netzer, Y.; Wang, T.; Coates, A.; Bissacco, A.; Wu, B.; Ng, A.Y. Reading digits in natural images with unsupervised feature learning. In Proceedings of the NIPS Workshop on Deep Learning and Unsupervised Feature Learning, Granada, Spain, 12–17 December 2011; pp. 1–9. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Malmström, E.; Kilsgård, O.; Hauri, S.; Smeds, E.; Herwald, H.; Malmström, L.; Malmström, J. Large-scale inference of protein tissue origin in gram-positive sepsis plasma using quantitative targeted proteomics. Nature Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Shelhamer, E.; Donahue, J.; Karayev, S.; Long, J.; Girshick, R.; Guadarrama, S.; Darrell, T. Caffe: Convolutional architecture for fast feature embedding. In Proceedings of the ACM International Conference on Multimedia, Orlando, FL, USA, 3–7 November 2014; pp. 675–678. [Google Scholar]
- Le, Q.V. Building high-level features using large scale unsupervised learning. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing, Vancouver, BC, Canada, 26–31 May 2013; pp. 8595–8598. [Google Scholar]
- Li, F.-F.; Rob, F.; Pietro, P. Learning generative visual models from few training examples: An incremental bayesian approach tested on 101 object categories. Comput. Visi. Image Underst. 2007, 106, 59–70. [Google Scholar]
- Lasseck, M. Audio-based bird species identification with deep convolutional neural networks. In Proceedings of the Working Notes of Conference and Labs of the Evaluation Forum, Avignon, France, 10–14 September 2018; pp. 1–11. [Google Scholar]
- Ji, L.; Ren, Y.; Liu, G.; Pu, X. Training-based gradient lbp feature models for multiresolution texture classification. IEEE Trans. Cybern. 2018, 48, 2683–2696. [Google Scholar] [CrossRef] [PubMed]
- Nanni, L.; Ghidoni, S.; Brahnam, S. Handcrafted vs. non-handcrafted features for computer vision classification. Pattern Recognit. 2017, 71, 158–172. [Google Scholar] [CrossRef]





| (A) | ||||
| 400 | 800 | 1200 | 1500 | |
| [6] | 0.753 | 0.778 | 0.790 | 0.781 |
| [8] | 0.763 | 0.775 | 0.784 | 0.789 |
| [11] | 0.740 | 0.763 | 0.787 | 0.806 |
| [12] | 0.755 | 0.779 | 0.793 | 0.791 |
| [2] | 0.763 | 0.775 | 0.786 | 0.797 |
| Full | 0.724 | 0.752 | 0.750 | 0.753 |
| \Tuning | 0.750 | 0.738 | 0.735 | 0.734 |
| Ours | 0.779 | 0.791 | 0.793 | 0.799 |
| (B) | ||||
| 1000 | 5000 | 10,000 | ||
| [6] | 0.844 | 0.923 | 0.951 | |
| [8] | 0.773 | 0.875 | 0.887 | |
| [11] | 0.776 | 0.835 | 0.850 | |
| [12] | 0.861 | 0.926 | 0.946 | |
| [14] | 0.843 | 0.928 | 0.952 | |
| [2] | 0.887 | 0.928 | 0.932 | |
| Full | 0.854 | 0.926 | 0.945 | |
| \Tuning | 0.859 | 0.863 | 0.862 | |
| Ours | 0.893 | 0.936 | 0.938 | |
| # of Images | Type of Protocol |
|---|---|
| Change amount of protein | |
| Change concentration protocol | |
| Unprocessed | |
| Removal of only the top two abundant proteins | |
| Focus on the top two abundant proteins | |
| Focus on 14 abundant proteins | |
| Plasma sample instead of serum | |
| Removal of sugar chain | |
| Other protocols |
| PPV | NPV | MCC | F1 | ACC | |
|---|---|---|---|---|---|
| [6] | 0.962 | 0.944 | 0.879 | 0.912 | 0.949 |
| [8] | 0.931 | 0.957 | 0.879 | 0.915 | 0.949 |
| [11] | 1 | 0.932 | 0.880 | 0.909 | 0.949 |
| [12] | 0.931 | 0.957 | 0.879 | 0.915 | 0.949 |
| [2] | 0.931 | 0.957 | 0.879 | 0.915 | 0.949 |
| L-SVM | 0.871 | 0.956 | 0.833 | 0.885 | 0.929 |
| K-SVM | 0.931 | 0.957 | 0.879 | 0.915 | 0.949 |
| Full | 0.929 | 0.943 | 0.854 | 0.897 | 0.939 |
| \Tuning | 0.857 | 0.914 | 0.756 | 0.828 | 0.898 |
| Ours | 1 | 0.971 | 0.952 | 0.966 | 0.980 |
| PPV | NPV | MCC | F1 | ACC | |
|---|---|---|---|---|---|
| [2] | 0.878 | 0.985 | 0.885 | 0.921 | 0.949 |
| Full | 0.963 | 0.944 | 0.879 | 0.912 | 0.949 |
| Ours | 0.966 | 0.971 | 0.923 | 0.949 | 0.969 |
| PPV | NPV | MCC | F1 | ACC | |
|---|---|---|---|---|---|
| Caltech-101 | 0.923 | 0.917 | 0.804 | 0.857 | 0.918 |
| CIFAR-10 | 0.936 | 0.985 | 0.929 | 0.951 | 0.969 |
| MNIST | 0.936 | 0.985 | 0.929 | 0.951 | 0.969 |
| 2-DE images | 1 | 0.971 | 0.952 | 0.966 | 0.980 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawada, Y.; Sato, Y.; Nakada, T.; Yamaguchi, S.; Ujimoto, K.; Hayashi, N. Improvement in Classification Performance Based on Target Vector Modification for All-Transfer Deep Learning. Appl. Sci. 2019, 9, 128. https://doi.org/10.3390/app9010128
Sawada Y, Sato Y, Nakada T, Yamaguchi S, Ujimoto K, Hayashi N. Improvement in Classification Performance Based on Target Vector Modification for All-Transfer Deep Learning. Applied Sciences. 2019; 9(1):128. https://doi.org/10.3390/app9010128
Chicago/Turabian StyleSawada, Yoshihide, Yoshikuni Sato, Toru Nakada, Shunta Yamaguchi, Kei Ujimoto, and Nobuhiro Hayashi. 2019. "Improvement in Classification Performance Based on Target Vector Modification for All-Transfer Deep Learning" Applied Sciences 9, no. 1: 128. https://doi.org/10.3390/app9010128