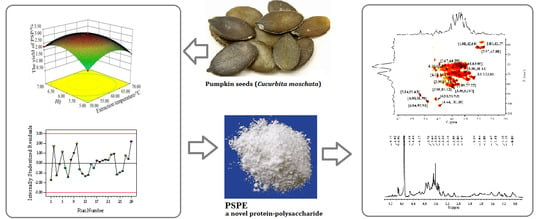
Enzyme Assisted Extraction, Purification and Structure Analysis of the Polysaccharides from Naked Pumpkin Seeds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Enzyme-Assisted Extraction of Polysaccharides
2.3. Optimization of EAE by RSM
2.4. Column Isolation of PSP
2.5. Characterization of PSPE
2.5.1. Determination of Molecular Weight and Monosaccharide Composition
2.5.2. UV and FT-IR Spectra
2.5.3. NMR Spectra
2.6. Statistical Analysis
3. Results and Discussion
3.1. Effects of Different Extraction Conditions on the Yield of PSP
3.1.1. Extraction Time
3.1.2. Extraction Temperature
3.1.3. Enzyme Concentration
3.1.4. The pH Values
3.2. Experimental Design and Analysis of Variance
3.2.1. Building Response Surface Model
3.2.2. Estimation of the Model Precision
3.2.3. Optimization of Experimental Procedures
3.2.4. Validation of the Predictive Model
3.3. Purification and Preliminary Characterisation of PSPE
3.4. FT-IR Spectrum Analysis
3.5. NMR Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Charoensiddhi, S.; Lorbeer, A.J.; Lahnstein, J.; Bulone, V.; Franco, C.M.M.; Zhang, W. Enzyme assisted extraction of carbohydrates from the brown alga Ecklonia radiata: Effect of enzyme type, pH and buffer on sugar yield and molecular weight profiles. Process Biochem. 2016, 51, 1503–1510. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, X.; Zhang, J. Optimization of enzyme assisted extraction of polysaccharides from Astragalus membranaceus. Carbohydr. Polym. 2014, 111, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Fernández, K.; Vega, M.; Aspé, E. An enzymatic extraction of proanthocyanidins from País grape seeds and skins. Food Chem. 2015, 168, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Babbar, N.; Van Roy, S.; Wijnants, M.; Dejonghe, W.; Caligiani, A.; Sforza, S.; Elst, K. Effect of extraction conditions on the saccharide (neutral and acidic) composition of the crude pectic extract from various agro-industrial residues. J. Agric. Food Chem. 2016, 64, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Mat Yusoff, M.; Gordon, M.H.; Ezeh, O.; Niranjan, K. Aqueous enzymatic extraction of Moringa oleifera oil. Food Chem. 2016, 211, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Feng, S.; Jia, X.; Li, Q.; Zhou, Y.; Ding, C. Structural characterization and antioxidant activities of polysaccharides extracted from Epimedium acuminatum. Carbohydr. Polym. 2013, 92, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, Y.; Li, W.; Wang, Z.; Shen, L. Optimization of polysaccharides extraction from Tricholoma mongolicum Imai and their antioxidant and antiproliferative activities. Carbohydr. Polym. 2015, 131, 322–330. [Google Scholar] [CrossRef] [PubMed]
- You, Q.; Yin, X.; Zhao, Y. Enzyme assisted extraction of polysaccharides from the fruit of Cornus officinalis. Carbohydr. Polym. 2013, 98, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Rostami, H.; Gharibzahedi, S.M.T. Cellulase-assisted extraction of polysaccharides from Malva sylvestris: Process optimization and potential functionalities. Int. J. Biol. Macromol. 2017, 101, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.M.; Song, J.H.; Wang, J.; Yang, J.M.; Wang, Z.B.; Liu, Y.H. Optimization of cellulase-assisted extraction process and antioxidant activities of polysaccharides from Tricholoma mongolicum Imai. J. Sci. Food Agric. 2016, 96, 4484–4491. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wu, D.; Ning, X.; Yang, G.; Lin, Z.; Tian, M.; Zhou, Y. α-Amylaseassisted extraction of polysaccharides from Panax ginseng. Int. J. Biol. Macromol. 2015, 75, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, M.; Guo, K.; Jia, A.; Shi, Y.; Gao, G.; Liu, C. Cellulase-assisted extraction, characterization, and bioactivity of polysaccharides from Polygonatum odoratum. Int. J. Biol. Macromol. 2015, 75, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.B.; Cheng, L.; Xu, Y.Q.; Wang, T.H.; Li, L.B.; Wang, H.S. Extraction technology optimization and antioxidant activity of polysaccharides from pumpkin seeds. Trans. Chin. Soc. Agric. Eng. 2016, 32, 284–290. [Google Scholar] [CrossRef]
- Wang, L.B.; Cheng, L.; Liu, F.C.; Li, T.F.; Yu, Z.Y.; Xu, Y.Q.; Yang, Y. Optimization of ultrasound-assisted extraction and structural characterization of the polysaccharide from pumpkin (Cucurbita moschata) seeds. Molecules 2018, 23, 1207. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.B.; Liu, F.C.; Wang, A.X.; Yu, Z.Y.; Xu, Y.Q.; Yang, Y. Purification, characterization and bioactivity determination of a novel polysaccharide from pumpkin (Cucurbita moschata) seeds. Food Hydrocoll. 2017, 66, 357–364. [Google Scholar] [CrossRef]
- Zhao, C.; Li, X.; Miao, J.; Jing, S.; Li, X.; Huang, L.; Gao, W. The effect of different extraction techniques on property and bioactivity of polysaccharides from Dioscorea hemsleyi. Int. J. Biol. Macromol. 2017, 102, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wong, P.T.S.; Dutka, B.J. Determination of carbohydrate in lake sediment by a modified phenol-sulfuric acid method. Water Res. 1973, 7, 741–746. [Google Scholar] [CrossRef]
- Lott, J.A.; Stephan, V.A.; Pritchard, K.A. Evaluation of the Coomassie Brilliant Blue G-250 method for urinary protein. Clin. Chem. 1983, 29, 1946–1950. [Google Scholar] [PubMed]
- Yu, Z.Y.; Liu, L.; Xu, Y.Q.; Wang, L.B.; Teng, X.; Li, X.G.; Li, X.J.; Dai, J. Characterization and biological activities of a novel polysaccharide isolated from raspberry (Rubus idaeus L.) fruits. Carbohydr. Polym. 2015, 132, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.C.; Miao, S.; Wen, X.C.; Sun, Y.X. Optimization of polysaccharides (ABP) extraction from the fruiting bodies of Agaricus blazei Murill using responsesurface methodology (RSM). Carbohydr. Polym. 2009, 78, 704–709. [Google Scholar] [CrossRef]
- Yin, X.L.; You, Q.H.; Jiang, Z.H. Optimization of enzyme assisted extractionof polysaccharides from Tricholoma matsutake by response surface methodology. Carbohydr. Polym. 2011, 86, 1358–1364. [Google Scholar] [CrossRef]
- Wu, H.; Zhu, J.; Diao, W.; Wang, C. Ultrasound-assisted enzymatic extraction and antioxidant activity of polysaccharides from pumpkin (Cucurbita moschata). Carbohydr. Polym. 2014, 113, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Jahanbin, K.; Abbasian, A.; Ahang, M. Isolation, purification and structural characterization of a new water-soluble polysaccharide from Eremurus stenophyllus (boiss. & buhse) baker roots. Carbohydr. Polym. 2017, 178, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.B.; Xu, Y.Q.; Yu, Z.Y.; Wang, T.H.; Li, L.B. Ethanol fractional precipitation and ultrasonic modification of pumpkin polysaccharides. Trans. Chin. Soc. Agric. Mach. 2015, 46, 215–220. [Google Scholar] [CrossRef]
- Ji, X.; Liu, F.; Peng, Q.; Wang, M. Purification, structural characterization, and hypolipidemic effects of a neutral polysaccharide from Ziziphus Jujuba cv. Muzao. Food Chem. 2018, 245, 1124–1130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, W.; Zheng, Y.; Jin, L.; Yao, W.; Gao, X. SGP-2, an acidic polysaccharide from Sarcandra glabra, inhibits proliferation and migration of human osteosarcoma cells. Food Funct. 2014, 5, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dobruchowska, J.M.; Gerwig, G.J.; Dijkhuizen, L.; Kamerling, J.P. Structural investigation of water-soluble polysaccharides extracted from the fruit bodies of Coprinus comatus. Carbohydr. Polym. 2013, 91, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.K. NMR spectroscopy in the structure elucidation of oligosaccharides and glycosides. Phytochemistry 1992, 31, 3307–3330. [Google Scholar] [CrossRef]
- Bosco, M.; Miertus, S.; Dentini, M.; Segre, A.L. The structure of gellan in dilute aqueous solution. Biopolymers 2000, 54, 115–126. [Google Scholar] [CrossRef]
- Yan, J.K.; Wang, Y.Y.; Ma, H.L.; Wang, Z.B.; Pei, J.J. Structural characteristics and antioxidant activity in vivo of a polysaccharide isolated from Phellinus linteus mycelia. J. Taiwan Inst. Chem. E 2016, 65, 110–117. [Google Scholar] [CrossRef]
- Dang, Z.L.; Feng, D.M.; Liu, X.H.; Yang, T.; Guo, L.; Liang, J.; Hu, F.; Cui, F.; Feng, S. Structure and antioxidant activity study of sulfated acetamido polysaccharide from Radix Hedysari. Fitoterapia 2013, 89, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Tang, Z.; Zhang, X.; Zhong, Y.; Yao, S.; Wang, L.; Lin, C.; Luo, X. Chemical properties and antioxidant activity of a water-soluble polysaccharide from Dendrobium officinale. Int. J. Biol. Macromol. 2016, 89, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.D.; Nie, S.P.; Yin, J.Y.; Que, Z.Q.; Zhang, L.J.; Huang, X.J. Polysaccharide from leaf skin of Aloe barbadensis Miller: Part I. Extraction, fractionation, physicochemical properties and structural characterization. Food Hydrocoll. 2017, 73, 176–183. [Google Scholar] [CrossRef]
- Liu, W.; Liu, Y.; Zhu, R.; Yu, J.; Lu, W.; Pan, C.; Yao, W.; Gao, X. Structure characterization, chemical and enzymatic degradation, andchain conformation of an acidic polysaccharide from Lycium barbarum L. Carbohydr. Polym. 2016, 147, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Ruthes, A.C.; Komura, D.L.; Carbonero, E.R.; Sassaki, G.L.; Gorin, P.A.J.; Iacomini, M. Structural characterization of the uncommon polysaccharides obtained from Peltigera canina photobiont Nostoc muscorum. Carbohydr. Polym. 2010, 81, 29–34. [Google Scholar] [CrossRef]
- Xu, Y.; Guo, Y.; Gao, Y.; Niu, X.; Wang, L.; Li, X.; Chen, H.; Yu, Z.; Yang, Y. Seperation, characterization and inhibition on α-glucosidase, α-amylase and glycation of a polysaccharide from blackcurrant fruits. LWT Food Sci. Technol. 2018, 93, 16–23. [Google Scholar] [CrossRef]








| Factors | Level | ||
|---|---|---|---|
| −1 | 0 | 1 | |
| A (Extraction time/min) | 30 | 40 | 50 |
| B (Extraction temperature/°C) | 50 | 60 | 70 |
| C (Enzyme concentration/%) | 2.0 | 2.5 | 3.0 |
| D (pH) | 5.0 | 6.0 | 7.0 |
| Number | A (Extraction Time, min) | B (Extraction Temperature, °C) | C (Enzyme Concentration, %) | D (pH) | Yield of PSP (Y) % |
|---|---|---|---|---|---|
| 1 | 40 | 70 | 3.0 | 6.0 | 2.31 |
| 2 | 40 | 60 | 2.0 | 5.0 | 2.47 |
| 3 | 50 | 60 | 2.0 | 6.0 | 2.78 |
| 4 | 40 | 60 | 3.0 | 5.0 | 2.29 |
| 5 | 40 | 50 | 2.0 | 6.0 | 2.72 |
| 6 | 50 | 60 | 2.5 | 5.0 | 1.92 |
| 7 | 50 | 50 | 2.5 | 6.0 | 1.71 |
| 8 | 40 | 60 | 2.5 | 6.0 | 3.22 |
| 9 | 30 | 60 | 3.0 | 6.0 | 2.25 |
| 10 | 50 | 70 | 2.5 | 6.0 | 2.68 |
| 11 | 40 | 70 | 2.5 | 5.0 | 1.73 |
| 12 | 40 | 60 | 2.5 | 6.0 | 3.08 |
| 13 | 40 | 70 | 2.0 | 6.0 | 2.01 |
| 14 | 30 | 50 | 2.5 | 6.0 | 1.97 |
| 15 | 30 | 60 | 2.5 | 7.0 | 1.38 |
| 16 | 40 | 70 | 2.5 | 7.0 | 1.53 |
| 17 | 40 | 60 | 2.5 | 6.0 | 3.21 |
| 18 | 40 | 60 | 2.5 | 6.0 | 3.20 |
| 19 | 50 | 60 | 2.5 | 7.0 | 2.44 |
| 20 | 40 | 60 | 2.5 | 6.0 | 3.22 |
| 21 | 40 | 50 | 2.5 | 5.0 | 1.67 |
| 22 | 40 | 60 | 2.0 | 7.0 | 2.48 |
| 23 | 50 | 60 | 3.0 | 6.0 | 2.77 |
| 24 | 30 | 60 | 2.0 | 6.0 | 1.79 |
| 25 | 40 | 60 | 3.0 | 7.0 | 2.54 |
| 26 | 30 | 60 | 2.5 | 5.0 | 1.41 |
| 27 | 40 | 50 | 3.0 | 6.0 | 2.50 |
| 28 | 40 | 50 | 2.5 | 7.0 | 2.27 |
| 29 | 30 | 70 | 2.5 | 6.0 | 0.93 |
| Source | Sum of Squares | DF | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Model | 10.50 | 14 | 0.75 | 63.31 | ** |
| A | 1.74 | 1 | 1.74 | 146.90 | ** |
| B | 0.23 | 1 | 0.23 | 19.15 | ** |
| C | 0.014 | 1 | 0.014 | 1.18 | |
| D | 0.11 | 1 | 0.11 | 9.30 | ** |
| AB | 1.01 | 1 | 1.01 | 85.25 | ** |
| AC | 0.055 | 1 | 0.055 | 4.66 | * |
| AD | 0.076 | 1 | 0.076 | 6.38 | * |
| BC | 0.068 | 1 | 0.068 | 5.71 | * |
| BD | 0.16 | 1 | 0.16 | 13.50 | ** |
| CD | 0.014 | 1 | 0.014 | 1.22 | |
| A2 | 3.14 | 1 | 3.14 | 264.83 | ** |
| B2 | 3.14 | 1 | 3,14 | 264.83 | ** |
| C2 | 0.047 | 1 | 0.047 | 4.00 | |
| D2 | 3.03 | 1 | 3.03 | 255.39 | ** |
| Residual | 0.17 | 14 | 0.012 | ||
| Lack of fit | 0.15 | 10 | 0.015 | 4.23 | |
| Pure error | 0.014 | 4 | 3.58 × 10−3 | ||
| Cor total | 10.67 | 28 | |||
| C.V.% | 4.75% | ||||
| R2 | 0.9845 | ||||
| adj-R2 | 0.9161 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Liu, F.; Li, T.; Liu, D.; Xu, Y.; Yang, Y. Enzyme Assisted Extraction, Purification and Structure Analysis of the Polysaccharides from Naked Pumpkin Seeds. Appl. Sci. 2018, 8, 1866. https://doi.org/10.3390/app8101866
Wang L, Liu F, Li T, Liu D, Xu Y, Yang Y. Enzyme Assisted Extraction, Purification and Structure Analysis of the Polysaccharides from Naked Pumpkin Seeds. Applied Sciences. 2018; 8(10):1866. https://doi.org/10.3390/app8101866
Chicago/Turabian StyleWang, Libo, Fangcheng Liu, Tengfei Li, Dawei Liu, Yaqin Xu, and Yu Yang. 2018. "Enzyme Assisted Extraction, Purification and Structure Analysis of the Polysaccharides from Naked Pumpkin Seeds" Applied Sciences 8, no. 10: 1866. https://doi.org/10.3390/app8101866