Short Communication: Obesity Intervention Resulting in Significant Changes in the Human Gut Viral Composition
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Description of Raw Data Sequences Used
3.2. Alterations of Gut Viral Diversity in Obesity and Post-Obesity Treatments
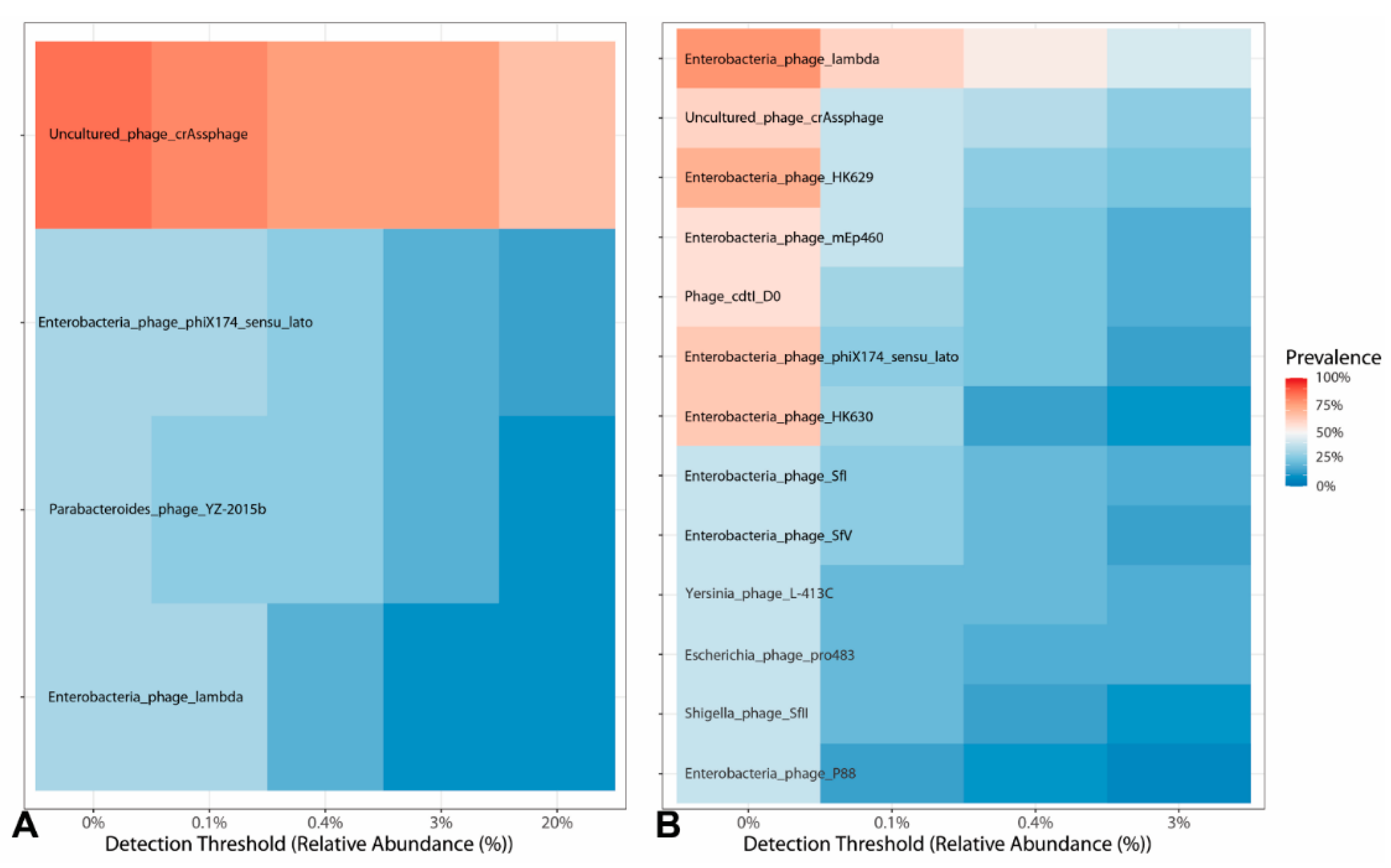
3.3. Viral Biomarker Enrichment in Each Condition
3.4. Core Virome in Pre- and Post-Obesity Intervention Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ellulu, M.; Abed, Y.; Rahmat, A.; Ranneh, Y.; Ali, F. Epidemiology of obesity in developing countries: Challenges and prevention. Glob. Epidemic Obes. 2014, 2, 2. [Google Scholar] [CrossRef] [Green Version]
- Pi-Sunyer, X. The medical risks of obesity. Postgrad. Med. 2009, 121, 21–33. [Google Scholar] [CrossRef]
- Wolfe, B.M.; Kvach, E.; Eckel, R.H. Treatment of Obesity: Weight Loss and Bariatric Surgery. Circ. Res. 2016, 118, 1844–1855. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Le, Q.A. Effectiveness of bariatric surgical procedures: A systematic review and network meta-analysis of randomized controlled trials. Medicine 2017, 96, 12–14. [Google Scholar] [CrossRef]
- Tevis, S.; Garren, M.J.; Gould, J.C. Revisional surgery for failed vertical-banded gastroplasty. Obes. Surg. 2011, 21, 1220–1224. [Google Scholar] [CrossRef]
- Andoh, A. Physiological role of gut microbiota for maintaining human health. Digestion 2016, 93, 176–181. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8836–8847. [Google Scholar] [CrossRef]
- Kho, Z.Y.; Lal, S.K. The human gut microbiome—A potential controller of wellness and disease. Front. Microbiol. 2018, 9, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, C.D. The gut microbiome and its role in obesity. Nutr. Today 2016, 51, 167–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palleja, A.; Kashani, A.; Allin, K.H.; Nielsen, T.; Zhang, C.; Li, Y.; Brach, T.; Liang, S.; Feng, Q.; Jogensen, N.B.; et al. Roux-en-Y gastric bypass surgery of morbidly obese patients induces swift and persistent changes of the individual gut microbiota. Genome Med. 2016, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Tremaroli, V.; Karlsson, F.; Werling, M.; Ståhlman, M.; Kovatcheva-Datchary, P.; Olbers, T.; Fändriks, L.; Le Roux, C.W.; Nielsen, J.; Bäckhed, F. Roux-en-Y Gastric Bypass and Vertical Banded Gastroplasty Induce Long-Term Changes on the Human Gut Microbiome Contributing to Fat Mass Regulation. Cell Metab. 2015, 22, 228–238. [Google Scholar] [CrossRef] [Green Version]
- Medina, D.A.; Li, T.; Thomson, P.; Artacho, A.; Pérez-Brocal, V.; Moya, A. Cross-regional view of functional and taxonomic microbiota composition in obesity and post-obesity treatment shows country specific microbial contribution. Front. Microbiol. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farin, W.; Oñate, F.P.; Plassais, J.; Bonny, C.; Beglinger, C.; Woelnerhanssen, B.; Nocca, D.; Magoules, F.; Le Chatelier, E.; Pons, N.; et al. Impact of laparoscopic Roux-en-Y gastric bypass and sleeve gastrectomy on gut microbiota: A metagenomic comparative analysis. Surg. Obes. Relat. Dis. 2020, 16, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Dao, M.C.; Everard, A.; Aron-Wisnewsky, J.; Sokolovska, N.; Prifti, E.; Verger, E.O.; Kayser, B.D.; Levenez, F.; Chilloux, J.; Hoyles, L.; et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut 2016, 65, 426–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabana; Shahid, S.U.; Irfan, U. The gut microbiota and its potential role in obesity. Future Microbiol. 2018, 13, 589–603. [Google Scholar] [CrossRef]
- Zhang, T.; Breitbart, M.; Lee, W.H.; Run, J.Q.; Wei, C.L.; Soh, S.W.L.; Hibberd, M.L.; Liu, E.T.; Rohwer, F.; Ruan, Y. RNA viral community in human feces: Prevalence of plant pathogenic viruses. PLoS Biol. 2006, 4, 0108–0118. [Google Scholar] [CrossRef] [Green Version]
- Carding, S.R.; Davis, N.; Hoyles, L.J.A.P. The human intestinal virome in health and disease. Aliment. Pharmacol. Ther. 2017, 46, 800–815. [Google Scholar] [CrossRef]
- Zuo, T.; Sun, Y.; Wan, Y.; Yeoh, Y.K.; Zhang, F.; Cheung, C.P.; Chen, N.; Luo, J.; Wang, W.; Sung, J.J.Y.; et al. Human-Gut-DNA Virome Variations across Geography, Ethnicity, and Urbanization. Cell Host Microbe 2020, 28, 741–751.e4. [Google Scholar] [CrossRef]
- Nayfach, S.; Páez-Espino, D.; Call, L.; Low, S.J.; Sberro, H.; Ivanova, N.N.; Proal, A.D.; Fischbach, M.A.; Bhatt, A.S.; Hugenholtz, P.; et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat. Microbiol. 2021, 6, 960–970. [Google Scholar] [CrossRef] [PubMed]
- Tithi, S.S.; Aylward, F.O.; Jensen, R.V.; Zhang, L. FastViromeExplorer: A pipeline for virus and phage identification and abundance profiling in metagenomics data. PeerJ 2018, 2018, 1–18. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org/ (accessed on 19 March 2021).
- Oksanen, A.J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Mcglinn, D.; Minchin, P.R.; Hara, R.B.O.; Simpson, G.L.; Solymos, P.; et al. CRAN-Package Vegan, Version 2.5-7. 2020. Available online: R-Project.org (accessed on 21 March 2021).
- Leo, L.; Sudarshan, S.; Ernst, F.G.M.; Huang, R.; Borman, T.; Braccia, D.C.; Corrada Bravo, H. Tools for microbiome analysis in R. 2017. Available online: http://microbiome.github.com/microbiome (accessed on 9 April 2021).
- Shetty, S.A.; Hugenholtz, F.; Lahti, L.; Smidt, H.; de Vos, W.M. Intestinal microbiome landscaping: Insight in community assemblage and implications for microbial modulation strategies. FEMS Microbiol. Rev. 2017, 41, 182–199. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Paradis, E.; Schliep, K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Ginestet, C. Ggplot2: Elegant graphics for data analysis. J.-R. Stat. Soc. Ser. A 2011, 174, 245. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Tremaroli, V.; Backhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, F.; Tremaroli, V.; Nielsen, J.; Backhed, F. Assessing the human gut microbiota in metabolic diseases. Diabetes Metab. Res. Rev. 2013, 62, 3341–3349. [Google Scholar] [CrossRef] [Green Version]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Mathieu, A.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Damms-Machado, A.; Mitra, S.; Schollenberger, A.E.; Kramer, K.M.; Meile, T.; Königsrainer, A.; Huson, D.H.; Bischoff, S.C. Effects of surgical and dietary weight loss therapy for obesity on gut microbiota composition and nutrient absorption. BioMed Res. Int. 2015, 2015, 806248. [Google Scholar] [CrossRef]
- Liang, G.; Bushman, F.D. The human virome: Assembly, composition and host interactions. Nat. Rev. Microbiol. 2021, 19, 514–527. [Google Scholar] [CrossRef]
- Galtier, M.; Sordi, L.D.; Sivignon, A.; De Vallée, A.; Maura, D.; Neut, C.; Rahmouni, O.; Wannerberger, K.; Darfeuille-Michaud, A.; Desreumaux, P.; et al. Bacteriophages targeting adherent invasive Escherichia coli strains as a promising new treatment for Crohn’s disease. J. Crohn’s Colitis 2017, 11, 840–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barr, J.J.; Auro, R.; Furlan, M.; Whiteson, K.L.; Erb, M.L.; Pogliano, J.; Stotland, A.; Wolkowicz, R.; Cutting, A.S.; Doran, K.S.; et al. Bacteriophage adhering to mucus provide a non–host-derived immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 10771–10776. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Niu, J.; Zuo, T.; Sun, Y.; Xu, Z.; Tang, W.; Liu, Q.; Zhang, J.; Ng, E.K.; Wong, S.K.; et al. Alterations in the gut virome in obesity and type 2 diabetes mellitus. Gastroenterology 2021, 161, 1257–1269. [Google Scholar] [CrossRef]
- Simmonds, P.; Adams, M.J.; Benkő, M.; Breitbart, M.; Brister, J.R.; Carstens, E.B.; Zerbini, F.M. Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017, 15, 161–168. [Google Scholar] [CrossRef]
- Jang, H.B.; Bolduc, B.; Zablocki, O.; Kuhn, J.H.; Roux, S.; Adriaenssens, E.M.; Sullivan, M.B. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 2019, 37, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Siddell, S.G.; Walker, P.J.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; et al. Additional changes to taxonomy ratified in a special vote by the International Committee on Taxonomy of Viruses (October 2018). Arch. Virol. 2019, 164, 943–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sandoval-Vargas, D.; Concha-Rubio, N.D.; Navarrete, P.; Castro, M.; Medina, D.A. Short Communication: Obesity Intervention Resulting in Significant Changes in the Human Gut Viral Composition. Appl. Sci. 2021, 11, 10039. https://doi.org/10.3390/app112110039
Sandoval-Vargas D, Concha-Rubio ND, Navarrete P, Castro M, Medina DA. Short Communication: Obesity Intervention Resulting in Significant Changes in the Human Gut Viral Composition. Applied Sciences. 2021; 11(21):10039. https://doi.org/10.3390/app112110039
Chicago/Turabian StyleSandoval-Vargas, Diego, Nataly D. Concha-Rubio, Paola Navarrete, Matías Castro, and Daniel A. Medina. 2021. "Short Communication: Obesity Intervention Resulting in Significant Changes in the Human Gut Viral Composition" Applied Sciences 11, no. 21: 10039. https://doi.org/10.3390/app112110039
APA StyleSandoval-Vargas, D., Concha-Rubio, N. D., Navarrete, P., Castro, M., & Medina, D. A. (2021). Short Communication: Obesity Intervention Resulting in Significant Changes in the Human Gut Viral Composition. Applied Sciences, 11(21), 10039. https://doi.org/10.3390/app112110039







