Active Participatory Regional Surveillance for Notifiable Swine Pathogens
Abstract
:Simple Summary
Abstract
1. Introduction
2. Study Design
2.1. Phase 1: Simulating Pathogen Spread—Animal Disease Spread Model (ADSM)
2.1.1. ADSM Simulations
2.1.2. ADSM Automation
- A new file directory in the ADSM workspace was created using the base R function dir.create and the saved template scenario “ScenarioX.db” was copied from the initial simulation. This file was renamed using the base R function file.rename, e.g., “new_scenario.db”.
- A connection was created between R and the SQLite database using the dbConnect function from the RSQLite R package (version 2.2.4) [30] to access the “new_scenario.db” file for editing, (i.e., con = dbConnect(SQLite(), dbname = “new_scenario.db”)).
- Once the connection was opened, the dbGetQuery function was used to bring the SQLite table to be edited into the R environment as a data frame. For example, the R code created a data frame in the R environment named “Population” using the ScenarioCreator_unit table from the SQLite database. This table contained the entire population file input during the ADSM scenario creation process.
- a.
- Population <- dbGetQuery(con, “SELECT * FROM ScenarioCreator_unit”).
- b.
- Other tables altered using this procedure included those containing the direct spread parameters (ScenarioCreator_directspread), indirect spread parameters (ScenarioCreator_indirectspread), and local area spread parameters (ScenarioCreator_airbornespread).
- R functions were then used to update the data frame to fit the new desired scenario (i.e., production types, initial infection statuses, or transmission probabilities).
- The dbWriteTable function with the overwrite option specified as TRUE was used to replace the SQLite table in the database file with the newly edited data frame. For example, dbWriteTable(con, name = “ScenarioCreator_unit”, value = Population, overwrite = TRUE).
- Rerunning the dbConnect line exactly as written in Step 2 saved the SQLite database file with the changes included (i.e., con = dbConnect(SQLite(), dbname = “new_scenarioX.db”)).
2.1.3. Phase 1: Spread Results
2.2. Phase 2: Simulating Pathogen Detection
- Assign as participants the ith set of participating farms from the list of pre-selected sets corresponding to the current setting’s participation level. For 100% participation, the entire population of farms were participants.
- For each participating farm, the farm infection status (negative, positive) was identified for days 7, 14, 21, 28, 35, 42, 49, 63, and 70 of the ADSM spread simulation.
- For each of the days listed in Step 2, participating farms classified as positive were “tested” (simulated) independently in R using the rbinom function with the probability of detection equaling the assigned farm-level sensitivity. Thus, for each positive farm and where p was the assigned farm-level sensitivity, rbinom (n = 1, size = 1, prob = p) randomly generated a 0 or 1, where 1 indicated that the infection was detected.
2.2.1. Phase 2: Detection Results
2.3. Phase 3: Cost of Sampling and Testing
2.3.1. Phase 3: Estimated Cost of Sampling and Testing
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- United States Department of Agriculture. Changes in the U.S. Swine Industry, 1995–2012 (#678.0817); USDA:APHIS:VS: CEAH, National Animal Health Monitoring System: Fort Collins, CO, USA, 2017.
- Dunn, J.W. The evolution of the U.S. swine industry. In Changes in the Live Pig Market in Different Countries; Szymańska, E.J., Ed.; Warsaw University of Life Sciences Press: Warsaw, Poland, 2017; pp. 19–27. [Google Scholar]
- Food and Agriculture Organization of the United Nations. FAOSTAT Statistical Database. Available online: www.fao.org/faostat/en/#data (accessed on 12 July 2021).
- United States Department of Agriculture. Meat Animals Production, Disposition, and Income—2020 Summary; USDA National Agricultural Statistics Service: Washington, DC, USA, 2021.
- Stevenson, G.W.; Hoang, H.; Schwartz, K.J.; Burrough, E.R.; Sun, D.; Madson, D.; Cooper, V.L.; Pillatzki, A.; Gauger, P.; Schmitt, B.J.; et al. Emergence of porcine epidemic diarrhea virus in the United States: Clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Investig. 2013, 25, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Niederwerder, M.C.; Hesse, R.A. Swine enteric coronavirus disease: A review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound. Emerg. Dis. 2018, 65, 660–675. [Google Scholar] [CrossRef] [PubMed]
- Moura, J.A.; McManus, C.M.; Bernal, F.E.M.; de Melo, C.B. An analysis of the 1978 African swine fever outbreak in Brazil and its eradication. Rev. Sci. Tech. Off. Int. Epiz. 2010, 29, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Stegeman, A.; Elbers, A.; de Smit, H.; Moser, H.; Smak, J.; Pluimers, F. The 1997–1998 epidemic of classical swine fever in the Netherlands. Vet. Microbiol. 2000, 73, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, M.P.; Horst, S.H.; Huirne, R.B.; Dijkhuizen, A.A. A model to estimate the financial consequences of classical swine fever outbreaks: Principles and outcomes. Prev. Vet. Med. 1999, 42, 249–270. [Google Scholar] [CrossRef]
- Davies, G. The foot and mouth disease (FMD) epidemic in the United Kingdom 2001. Comp. Immunol. Microbiol. Infect. Dis. 2002, 25, 331–343. [Google Scholar] [CrossRef]
- World Organisation for Animal Health. African Swine Fever (ASF)—Situation Report 9 (7 April 2022). Available online: https://www.oie.int/en/what-we-do/animal-health-and-welfare/disease-data-collection/ (accessed on 21 April 2022).
- Doherr, M.G.; Audigé, L. Monitoring and surveillance for rare health-related events: A review from the veterinary perspective. Phil. Trans. R. Soc. Lond. B 2001, 356, 1097–1106. [Google Scholar] [CrossRef]
- Anderson, L.A.; Black, N.; Hagerty, T.J.; Kluge, J.P.; Sundberg, P.L. Pseudorabies (Aujeszky’s Disease) and Its Eradication. A Review of the U.S. Experience; Technical Bulletin No. 1923; United States Department of Agriculture, Animal and Plant Health Inspection Service: Riverdale, MD, USA, 2008.
- Hoinville, L.J.; Alban, L.; Dewe, J.A.; Gibbens, J.C.; Gustafson, L.; Häsler, B.; Saegerman, C.; Salman, M.; Stärk, K.D.C. Proposed terms and concepts for describing and evaluating animal-health surveillance systems. Prev. Vet. Med. 2013, 112, 1–12. [Google Scholar] [CrossRef]
- Bush, E.J.; Gardner, I.A. Animal health surveillance in the United States via the National Animal Health Monitoring System (NAHMS). Epidemiol. Sante Anim. 1995, 27, 113–126. [Google Scholar]
- Gates, M.C.; Earl, L.; Enticott, G. Factors influencing the performance of voluntary farmer disease reporting in passive surveillance systems: A scoping review. Prev. Vet. Med. 2021, 196, 105487. [Google Scholar] [CrossRef] [PubMed]
- Mariner, J.C.; Hendrickx, S.; Pfeiffer, D.U.; Costard, S.; Knopf, L.; Okuthe, S.; Chibeu, D.; Parmley, J.; Musenero, M.; Pisang, C.; et al. Integration of participatory approaches into surveillance systems. Rev. Sci. Tech. Off. Int. Epiz. 2011, 30, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Calba, C.; Antoine-Moussiaux, N.; Charrier, F.; Hendrikx, P.; Saegerman, C.; Peyre, M.; Goutard, F.L. Applying participatory approaches in the evaluation of surveillance systems: A pilot study on African swine fever surveillance in Corsica. Prev. Vet. Med. 2015, 122, 389–398. [Google Scholar] [CrossRef]
- Wójcik, O.P.; Brownstein, J.S.; Chunara, R.; Johansson, M.A. Public health for the people: Participatory infectious disease surveillance in the digital age. Emerg. Themes Epidemiol. 2014, 11, 7. [Google Scholar] [CrossRef]
- Smolinski, M.S.; Crawley, A.W.; Olsen, J.M.; Jayaraman, T.; Libel, M. Participatory disease surveillance: Engaging communities directly in reporting, monitoring, and responding to health threats. JMIR Pub. Health Suveill. 2017, 3, e7540. [Google Scholar] [CrossRef] [PubMed]
- Elbers, A.R.W.; Bouma, A.; Stegeman, J.A. Quantitative assessment of clinical signs for the detection of classical swine fever outbreaks during an epidemic. Vet. Microbiol. 2002, 85, 323–332. [Google Scholar] [CrossRef]
- Harvey, N.; Reeves, A.; Schoenbaum, M.A.; Zagmutt-Vergara, F.J.; Dubé, C.; Hill, A.E.; Corso, B.A.; McNab, W.B.; Cartwright, C.I.; Salman, M.D. The North American Animal Disease Spread Model: A simulation model to assist decision making in evaluating animal disease incursions. Prev. Vet. Med. 2007, 82, 176–197. [Google Scholar] [CrossRef]
- Lowe, J.; Gauger, P.; Harmon, K.; Zhang, J.; Connor, J.; Yeske, P.; Loula, T.; Levis, I.; Dufresne, L.; Main, R. Role of transportation in the spread of porcine epidemic diarrhea virus infection, United States. Emerg. Infect. Dis. 2014, 20, 872–874. [Google Scholar] [CrossRef]
- Meyer, S. Pork Packing: Just What Is Capacity? National Hog Farmer, 6 October 2020; pp. 18–22. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org (accessed on 30 May 2021).
- Hagenstein, P.R.; Flocchini, R.G.; Bailar, J.C., III; Claiborn, C.; Dickerson, R.R.; Galloway, J.N.; Grossman, M.R.; Kasibhatla, P.; Kohn, R.A.; Lacy, M.P.; et al. The Scientific Basis for Estimating Air Emissions from Animal Feeding Operations: Interim Report; The National Academy Press: Washington, DC, USA, 2002; p. 90. [Google Scholar]
- Bivand, R.S.; Pebesma, E.; Gómez-Rubio, V. Applied Spatial Data Analysis with R, 2nd ed.; Springer: New York, NY, USA, 2013; pp. 146–149. [Google Scholar]
- Thakur, K.K.; Revie, C.W.; Hurnik, D.; Poljak, Z.; Sanchez, J. Simulation of between-farm transmission of porcine reproductive and respiratory syndrome virus in Ontario, Canada using the North American Animal Disease Spread Model. Prev. Vet. Med. 2015, 118, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Machado, G.; Galvis, J.A.; Lopes, F.P.N.; Voges, J.; Medeiros, A.A.R.; Cárdenas, N.C. Quantifying the dynamics of pig movements improves targeted disease surveillance and control plans. Transbound. Emerg. Dis. 2021, 68, 1663–1675. [Google Scholar] [CrossRef]
- Müller, K.; Wickham, H.; James, D.A.; Falcon, S. RSQLite: SQLite Interface for R, Version 2.2.4. Available online: https://CRAN.R-project.org/package=RSQLite (accessed on 12 June 2022).
- Thacker, S.B.; Parrish, R.G.; Trowbridge, F.L. A method for evaluating systems of epidemiological surveillance. World Health Stat. Q. 1988, 41, 11–18. [Google Scholar]
- Nielsen, S.S.; Alvarez, J.; Bicout, D.J.; Calistri, P.; Canali, E.; Drewe, J.A.; Garin-Bastuji, B.; Gonzales Rojas, J.L.; Schmidt, C.G.; Herskin, M.; et al. Assessment of the control measures of the category A diseases of animal health law: Classical swine fever. EFSA J. 2021, 19, e06707. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.S.; Alvarez, J.; Bicout, D.J.; Calistri, P.; Depner, K.; Drewe, J.A.; Garin-Bastuji, B.; Gonzales Rojas, J.L.; Schmidt, C.G.; Herskin, M.; et al. Scientific opinion on the assessment of the control measures of the category A diseases of the animal health law: African swine fever. EFSA J. 2021, 19, e06402. [Google Scholar] [CrossRef]
- Christensen, J.; Gardner, I.A. Herd-level interpretation of test results for epidemiologic studies of animal diseases. Prev. Vet. Med. 2000, 45, 83–106. [Google Scholar] [CrossRef] [PubMed]
- Crauwels, A.P.P.; Nielen, M.; Stegeman, J.A.; Elbers, A.R.W.; Dijkhuizen, A.A.; Tielen, M.J.M. The effectiveness of routine serological surveillance: Case study of the 1997 epidemic of classical swine fever in the Netherlands. Rev. Sci. Tech. Off. Int. Epiz. 1999, 18, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, P.D.; Le Potier, M.-F.; Finlaison, D. Pestiviruses. In Diseases of Swine, 11th ed.; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2019; pp. 622–640. [Google Scholar] [CrossRef]
- Gallardo, C.; Soler, A.; Rodze, I.; Nieto, R.; Cano-Gómez, C.; Fernandez-Pinero, J.; Arias, M. Attenuated and non-haemadsorbing (non-HAD) genotype II African swine fever virus (ASFV) isolated in Europe, Latvia 2017. Transbound. Emerg. Dis. 2019, 66, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Bates, T.W.; Thurmond, M.C.; Hietala, S.K.; Venkateswaran, K.S.; Wilson, T.M.; Colston, B.W., Jr.; Trebes, J.E.; Milanovich, F.P. Surveillance for detection of foot-and-mouth disease. J. Am. Vet. Med. Assoc. 2003, 223, 609–614. [Google Scholar] [CrossRef]
- Schulz, K.; Conraths, F.J.; Blome, S.; Staubach, C.; Sauter-Louis, C. African swine fever: Fast and furious or slow and steady? Viruses 2019, 11, 866. [Google Scholar] [CrossRef] [PubMed]
- Munguía-Ramírez, B.; Armenta-Leyva, B.; Giménez-Lirola, L.; Wang, C.; Zimmerman, J. Surveillance on swine farms using antemortem specimens. In Optimising Pig Herd Health and Production; Maes, D., Segalés, J., Eds.; Burleigh Dodds Science Publishing: Cambridge, UK, 2023; pp. 97–138. [Google Scholar]
- Trevisan, G.; Linhares, L.C.M.; Crim, B.; Dubey, P.; Schwartz, K.J.; Burrough, E.R.; Main, R.G.; Sundberg, P.; Thurn, M.; Lages, P.T.F.; et al. Macroepidemiological aspects of porcine reproductive and respiratory syndrome virus detection by major United States veterinary diagnostic laboratories over time, age group, and specimen. PLoS ONE 2019, 10, e0223544. [Google Scholar] [CrossRef] [PubMed]
- Branson, B.M. Home sample collection tests for HIV infection. J. Am. Med. Assoc. 1998, 280, 1699–1701. [Google Scholar] [CrossRef]
- Tsang, N.N.Y.; So, H.C.; Ng, K.Y.; Cowling, B.J.; Leung, G.M.; Ip, D.K.M. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: A systematic review and meta-analysis. Lancet Infect. Dis. 2021, 21, 1233–1245. [Google Scholar] [CrossRef] [PubMed]
- Fatima, M.; Luo, Y.; Zhang, L.; Wang, P.-Y.; Song, H.; Fu, Y.; Li, Y.; Sun, Y.; Li, S.; Bao, Y.-J.; et al. Genotyping and molecular characterization of classical swine fever virus isolated in China during 2016–2018. Viruses 2021, 13, 664. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, G.; Frost, J.W. Study on the suitability of sow colostrum for the serological diagnosis of porcine reproductive and respiratory syndrome (PRRS). J. Vet. Med. B 1997, 44, 65–72. [Google Scholar] [CrossRef]
- Nijsten, R.; London, N.; van den Bogaard, A.; Stobberingh, E. Antibiotic resistance among Escherichia coli isolated from faecal samples of pig farmers and pigs. J. Antimicrob. Chemother. 1996, 37, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Newberry, K.M.; Colling, A. Quality standards and guidelines for test validation for infectious diseases in veterinary laboratories. Rev. Sci. Tech. Off. Int. Epiz. 2021, 40, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro Miguel, A.L.; Lopes Moreira, R.P. Fernando de Oliveira. ISO/IEC 17025: History and introduction of concepts. Química Nova 2021, 44, 792–796. [Google Scholar] [CrossRef]
- Hobbs, E.C.; Colling, A.; Gurung, R.B.; Allen, J. The potential of diagnostic point-of-care tests (POCTs) for infectious and zoonotic animal diseases in developing countries: Technical, regulatory and sociocultural considerations. Transbound. Emerg. Dis. 2021, 68, 1835–1849. [Google Scholar] [CrossRef] [PubMed]
- U.S. Census Bureau. State Area Measurements and Internal Point Coordinates. 2010. Available online: https://www.census.gov/geographies/reference-files/2010/geo/state-area.html (accessed on 25 June 2023).
- United Nations. Statistical Yearbook, 65th ed.; United Nations: New York, NY, USA, 2022; pp. 13–35. [Google Scholar]
- Ketusing, N.; Reeves, A.; Portacci, K.; Yano, T.; Olea-Popelka, F.; Keefe, T.; Salman, M. Evaluation of strategies for the eradication of pseudorabies virus (Aujeszky’s disease) in commercial swine farms in Chiang-Mai and Lampoon provinces, Thailand, using a simulation disease spread model. Transbound. Emerg. Dis. 2014, 61, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Stanojevic, S.; Valcic, M.; Stanojevic, S.; Radojicic, S.; Avramov, S.; Tambur, Z. Simulation of a classical swine fever outbreak in rural areas of the Republic of Serbia. Vet. Med. 2015, 60, 553–566. [Google Scholar] [CrossRef]
- Hasahya, E.; Thakur, K.K.; Dione, M.M.; Wieland, B.; Oba, P.; Kungu, J.; Lee, H.S. Modeling the spread of porcine reproductive and respiratory syndrome among pig farms in Lira district of northern Uganda. Front. Vet. Sci. 2021, 8, 727895. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Schulz, L.L.; Tonsor, G.T. Swine producer willingness to pay for Tier 1 disease risk mitigation under multifaceted ambiguity. Agribusiness 2021, 37, 858–875. [Google Scholar] [CrossRef]
| Total Pig Inventory | Breeder Sites (No. Pigs) | Breeder–Feeder Sites (No. Pigs) | Feeder Sites (No. Pigs) | Total Sites (No. Pigs) |
|---|---|---|---|---|
| ≤1000 | 476 | 650 | 3296 | 4422 |
| (140,823) | (254,214) | (980,356) | (1,375,393) | |
| 1001 to 4999 | 474 | 294 | 10,493 | 11,261 |
| (1,244,232) | (641,930) | (29,374,884) | (31,261,046) | |
| ≥5000 | 132 | 120 | 1586 | 1838 |
| (1,556,655) | (1,595,964) | (15,726,642) | (18,879,261) | |
| TOTAL | 1082 | 1064 | 15,375 | 17,521 |
| (2,941,710) | (2,492,108) | (46,081,882) | (51,515,700) |
| Spread Parameters | Parameter Definitions and/or Values | |||
|---|---|---|---|---|
|
| |||
|
| |||
| Destination | ||||
| Source farm | Breeder [28] | Feeder [28] | Packing Plant [29] | |
| Breeder–Feeder | NA | 0.0204 | 0.0310 | |
| Breeder | 0.0014 | 0.0687 | 0.0310 | |
| Feeder | NA | 0.0348 | 0.0310 | |
|
| |||
|
| |||
| - - - - - - - - - Destination - - - - - - - - - | ||||
| Source farm | Breeder | Feeder | Packing Plant | |
| Breeder–Feeder | NA | 0.0204 | 0.0310 | |
| Breeder | 0.0014 | 0.0687 | 0.0310 | |
| Feeder | NA | 0.0348 | 0.0310 | |
|
| |||
|
| |||
| Index Farm Location and Type 2 | Spread Probabilities | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Area Spread | Direct Contact 0.2 | Direct Contact 0.4 | Direct Contact 0.6 | ||||||||
| Indirect Contact | Indirect Contact | Indirect Contact | |||||||||
| 0.05 | 0.10 | 0.15 | 0.05 | 0.10 | 0.15 | 0.05 | 0.10 | 0.15 | |||
| Low-density county 1.1–3.3 pigs per km2 | BF | 0.001 | 4 | 4 | 5 | 16 | 16 | 21 | 63 | 67 | 81 |
| B | 10 | 11 | 12 | 46 | 54 | 60 | 191 | 218 | 242 | ||
| F | 6 | 7 | 8 | 25 | 31 | 36 | 99 | 113 | 138 | ||
| BF | 0.010 | 5 | 5 | 5 | 19 | 22 | 25 | 86 | 96 | 107 | |
| B | 11 | 12 | 15 | 59 | 64 | 75 | 245 | 279 | 318 | ||
| F | 7 | 8 | 10 | 31 | 39 | 45 | 136 | 160 | 182 | ||
| BF | 0.100 | 13 | 14 | 15 | 85 | 97 | 119 | 430 | 427 | 534 | |
| B | 34 | 42 | 48 | 247 | 298 | 311 | 1127 | 1223 | 1366 | ||
| F | 19 | 25 | 33 | 135 | 164 | 190 | 637 | 754 | 864 | ||
| Medium-density county 15.9–25.3 pigs per km2 | BF | 0.001 | 4 | 4 | 4 | 16 | 17 | 19 | 64 | 76 | 79 |
| B | 10 | 11 | 12 | 47 | 55 | 62 | 195 | 218 | 250 | ||
| F | 6 | 7 | 9 | 27 | 31 | 37 | 99 | 126 | 150 | ||
| BF | 0.010 | 5 | 5 | 6 | 21 | 23 | 29 | 88 | 102 | 112 | |
| B | 12 | 13 | 15 | 64 | 67 | 80 | 260 | 304 | 336 | ||
| F | 7 | 10 | 10 | 35 | 41 | 49 | 141 | 164 | 187 | ||
| BF | 0.100 | 16 | 18 | 18 | 119 | 120 | 146 | 456 | 510 | 595 | |
| B | 42 | 49 | 58 | 292 | 317 | 393 | 1325 | 1396 | 1539 | ||
| F | 23 | 30 | 40 | 155 | 200 | 209 | 742 | 859 | 956 | ||
| High-density county 106.8–214.5 pigs per km2 | BF | 0.001 | 5 | 5 | 5 | 19 | 20 | 21 | 66 | 74 | 85 |
| B | 11 | 12 | 13 | 51 | 58 | 65 | 200 | 233 | 271 | ||
| F | 6 | 8 | 9 | 26 | 34 | 41 | 108 | 132 | 151 | ||
| BF | 0.010 | 6 | 7 | 8 | 26 | 31 | 33 | 109 | 123 | 141 | |
| B | 14 | 16 | 18 | 71 | 80 | 94 | 285 | 322 | 358 | ||
| F | 10 | 11 | 13 | 42 | 51 | 55 | 164 | 182 | 210 | ||
| BF | 0.100 | 47 | 56 | 67 | 244 | 286 | 330 | 982 | 1015 | 1288 | |
| B | 81 | 89 | 102 | 446 | 500 | 575 | 1661 | 1936 | 2093 | ||
| F | 57 | 72 | 85 | 310 | 368 | 432 | 1239 | 1345 | 1553 | ||
| Index Farm Location 1 | Spread Probabilities | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Area Spread | Direct Contact 0.2 | Direct Contact 0.4 | Direct Contact 0.6 | |||||||
| Indirect Contact | Indirect Contact | Indirect Contact | ||||||||
| 0.05 | 0.10 | 0.15 | 0.05 | 0.10 | 0.15 | 0.05 | 0.10 | 0.15 | ||
| Low-density county | 0.001 | - | - | - | - | - | - | - | - | - |
| 0.010 | - | ✓ | - | - | ✓ | - | - | ✓ | - | |
| 0.100 | - | - | - | - | - | - | - | - | - | |
| Medium-density county | 0.001 | - | - | - | - | - | - | - | - | - |
| 0.010 | - | ✓ | - | - | ✓ | - | - | ✓ | - | |
| 0.100 | - | - | - | - | - | - | - | - | - | |
| High-density county | 0.001 | - | - | - | - | - | - | - | - | - |
| 0.010 | - | ✓ | - | - | ✓ | - | - | ✓ | - | |
| 0.100 | - | ✓ | - | - | ✓ | - | - | ✓ | - | |
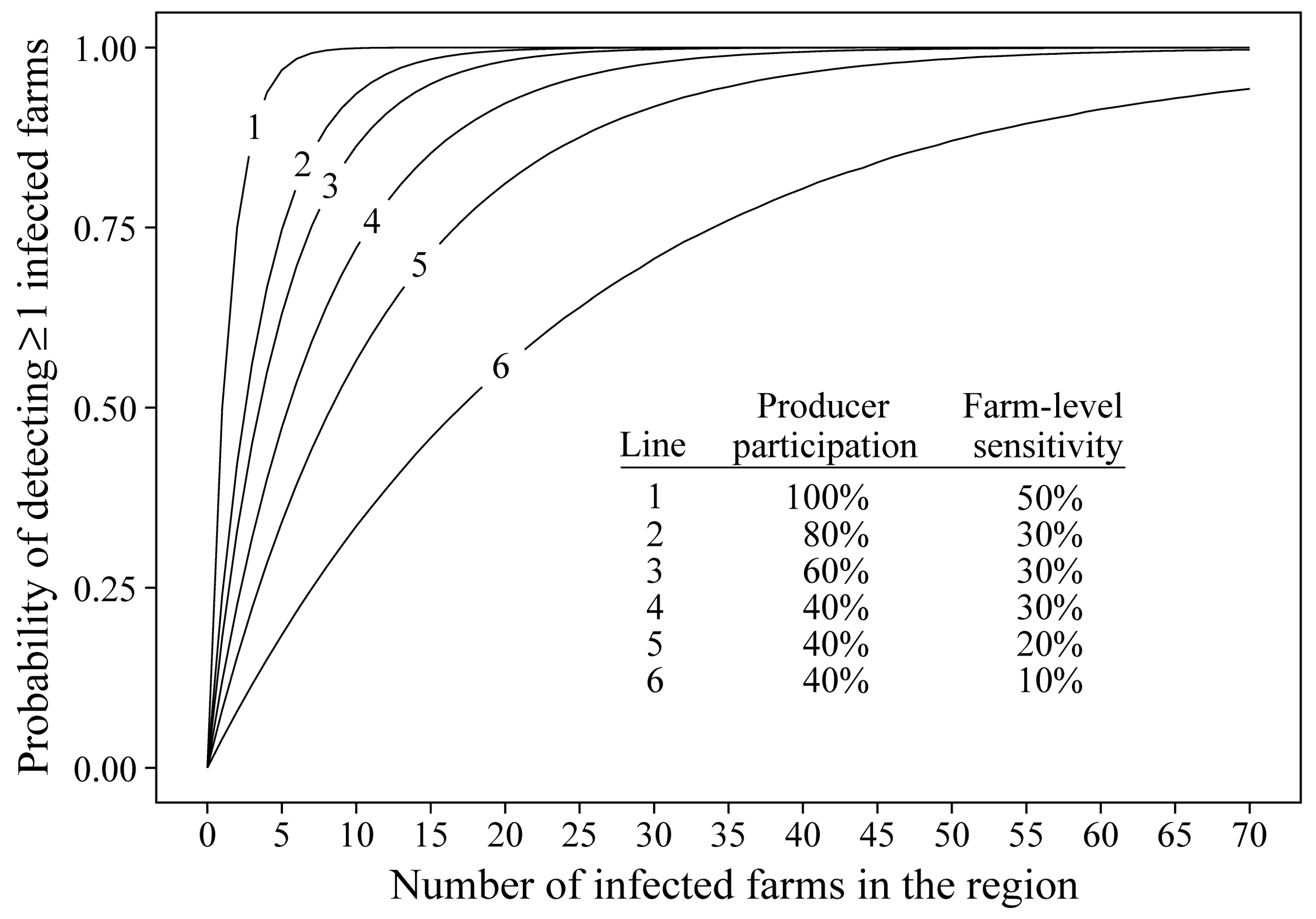
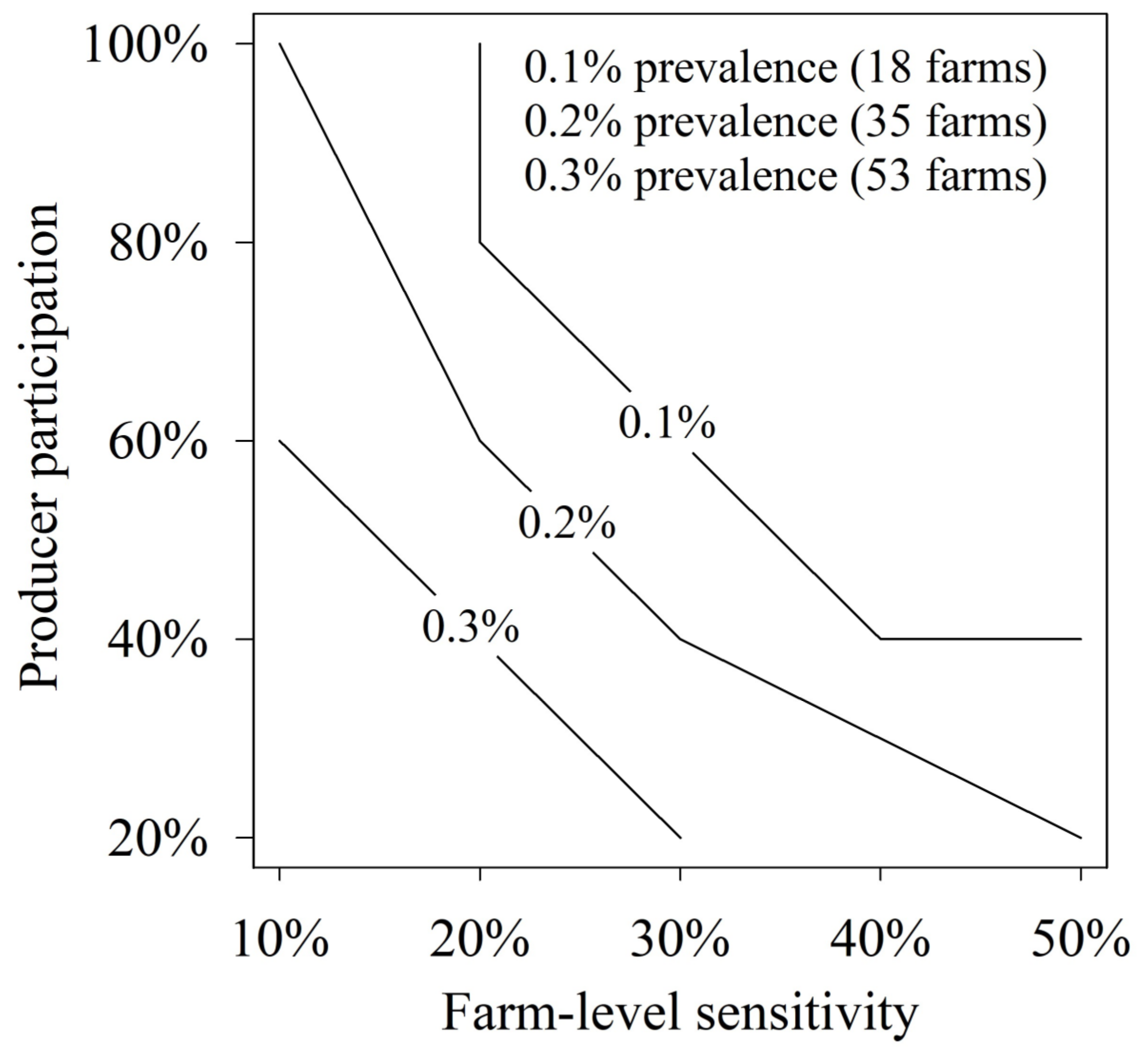
| Regional Prevalence 1 | Farm-Level Sensitivity (%) 2 | Producer Participation | ||||
|---|---|---|---|---|---|---|
| 20% | 40% | 60% | 80% | 100% | ||
| 0.1% (18 farms) | 10 | 0.304 | 0.520 | 0.672 | 0.777 | 0.850 |
| 20 | 0.519 | 0.777 | 0.900 | 0.957 | 0.982 | |
| 30 | 0.671 | 0.900 | 0.972 | 0.993 | 0.998 | |
| 40 | 0.776 | 0.956 | 0.993 | 0.999 | 1.000 | |
| 50 | 0.849 | 0.982 | 0.998 | 1.000 | 1.000 | |
| 0.2% (35 farms) | 10 | 0.506 | 0.760 | 0.886 | 0.946 | 0.975 |
| 20 | 0.760 | 0.945 | 0.989 | 0.998 | 1.000 | |
| 30 | 0.885 | 0.989 | 0.999 | 1.000 | 1.000 | |
| 40 | 0.945 | 0.998 | 1.000 | 1.000 | 1.000 | |
| 50 | 0.975 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 0.3% (53 farms) | 10 | 0.657 | 0.885 | 0.963 | 0.988 | 0.996 |
| 20 | 0.885 | 0.988 | 0.999 | 1.000 | 1.000 | |
| 30 | 0.962 | 0.999 | 1.000 | 1.000 | 1.000 | |
| 40 | 0.988 | 1.000 | 1.000 | 1.000 | 1.000 | |
| 50 | 0.996 | 1.000 | 1.000 | 1.000 | 1.000 | |
| Category | Cost per Item 2 | No. Items | Cost per Sampling | ||
|---|---|---|---|---|---|
| A. Sample collection. Assumes 10 pigs/farm/sampling | |||||
| Option 1. Serum samples | |||||
| Blood collection tubes (single-use) | 0.525 | 0.563 | 10 | 5.26 | 5.64 |
| Blood collection needles (single-use) | 0.572 | 0.613 | 10 | 5.72 | 6.13 |
| Plastic tube for pooling 5 samples 3 | 0.171 | 0.183 | 2 | 0.35 | 0.37 |
| Disposable gloves | €0.064 | $0.069 | 4 (2 pairs) | 0.26 | 0.28 |
| €11.58 | $12.42 | ||||
| Option 2. Swab samples (blood, nasal, oral, or fecal) | |||||
| Sample collection swabs | 0.532 | 0.570 | 10 | 5.32 | 5.70 |
| Transport medium, e.g., phosphate-buffered saline | 0.036 | 0.039 | 5 mL | 0.36 | 0.39 |
| Plastic tube for pooling 5 samples | 0.171 | 0.183 | 2 | 0.35 | 0.37 |
| Disposable gloves | €0.064 | $0.069 | 4 (2 pairs) | 0.26 | 0.28 |
| €6.29 | $6.74 | ||||
| B. Shipment of samples to the laboratory | |||||
| Insulated shipping container | 5.167 | 5.540 | 1 | 5.17 | 5.54 |
| Cold packs to ship with samples | 6.529 | 7.000 | 1 | 6.53 | 7.00 |
| Parcel shipping charge | €13.991 | $15.000 | 13.99 | 15.00 | |
| €25.69 | $27.54 | ||||
| Serum Tested by PCR | Swabs Tested by PCR | Serum Tested by ELISA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cost per test | EUR 18.65 | EUR 23.32 | EUR 27.98 | EUR 18.65 | EUR 23.32 | EUR 27.98 | EUR 4.66 | EUR 7.00 | EUR 9.33 | |
| Denominator | USD 20.00 | USD 25.00 | USD 30.00 | USD 20.00 | USD 25.00 | USD 30.00 | USD 5.00 | USD 7.50 | USD 10.00 | |
| Per farm in region 1 | EUR 74.56 | EUR 83.89 | EUR 93.22 | EUR 69.28 | EUR 78.60 | EUR 87.93 | EUR 46.58 | EUR 51.25 | EUR 55.91 | |
| USD 79.94 | USD 89.94 | USD 99.94 | USD 74.27 | USD 84.27 | USD 94.27 | USD 49.94 | USD 54.94 | USD 59.94 | ||
| Per pig in region 1 | EUR 0.025 | EUR 0.029 | EUR 0.032 | EUR 0.023 | EUR 0.027 | EUR 0.030 | EUR 0.016 | EUR 0.018 | EUR 0.019 | |
| USD 0.027 | USD 0.031 | USD 0.034 | USD 0.025 | USD 0.029 | USD 0.032 | USD 0.017 | USD 0.019 | USD 0.020 | ||
| Per pig in inventory Farms of ≤ 1000 pigs 2 | EUR 0.240 | EUR 0.270 | EUR 0.299 | EUR 0.223 | EUR 0.253 | EUR 0.283 | EUR 0.150 | EUR 0.165 | EUR 0.180 | |
| USD 0.257 | USD 0.289 | USD 0.321 | USD 0.239 | USD 0.271 | USD 0.303 | USD 0.161 | USD 0.177 | USD 0.193 | ||
| Farms of 1001–4999 pigs 3 | EUR 0.027 | EUR 0.030 | EUR 0.034 | EUR 0.025 | EUR 0.028 | EUR 0.032 | EUR 0.017 | EUR 0.019 | EUR 0.021 | |
| USD 0.029 | USD 0.032 | USD 0.036 | USD 0.027 | USD 0.030 | USD 0.034 | USD 0.018 | USD 0.020 | USD 0.022 | ||
| Farms of ≥ 5000 pigs 4 | EUR 0.007 | EUR 0.008 | EUR 0.009 | EUR 0.007 | EUR 0.007 | EUR 0.008 | EUR 0.005 | EUR 0.006 | EUR 0.006 | |
| USD 0.008 | USD 0.009 | USD 0.010 | USD 0.007 | USD 0.008 | USD 0.009 | USD 0.005 | USD 0.005 | USD 0.006 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trevisan, G.; Morris, P.; Silva, G.S.; Nakkirt, P.; Wang, C.; Main, R.; Zimmerman, J. Active Participatory Regional Surveillance for Notifiable Swine Pathogens. Animals 2024, 14, 233. https://doi.org/10.3390/ani14020233
Trevisan G, Morris P, Silva GS, Nakkirt P, Wang C, Main R, Zimmerman J. Active Participatory Regional Surveillance for Notifiable Swine Pathogens. Animals. 2024; 14(2):233. https://doi.org/10.3390/ani14020233
Chicago/Turabian StyleTrevisan, Giovani, Paul Morris, Gustavo S. Silva, Pormate Nakkirt, Chong Wang, Rodger Main, and Jeffrey Zimmerman. 2024. "Active Participatory Regional Surveillance for Notifiable Swine Pathogens" Animals 14, no. 2: 233. https://doi.org/10.3390/ani14020233





