Human Disturbance Increases Health Risks to Golden Snub-Nosed Monkeys and the Transfer Risk of Pathogenic Antibiotic-Resistant Bacteria from Golden Snub-Nosed Monkeys to Humans
Abstract
:Simple Summary
Abstract
1. Introduction
2. Research Methods
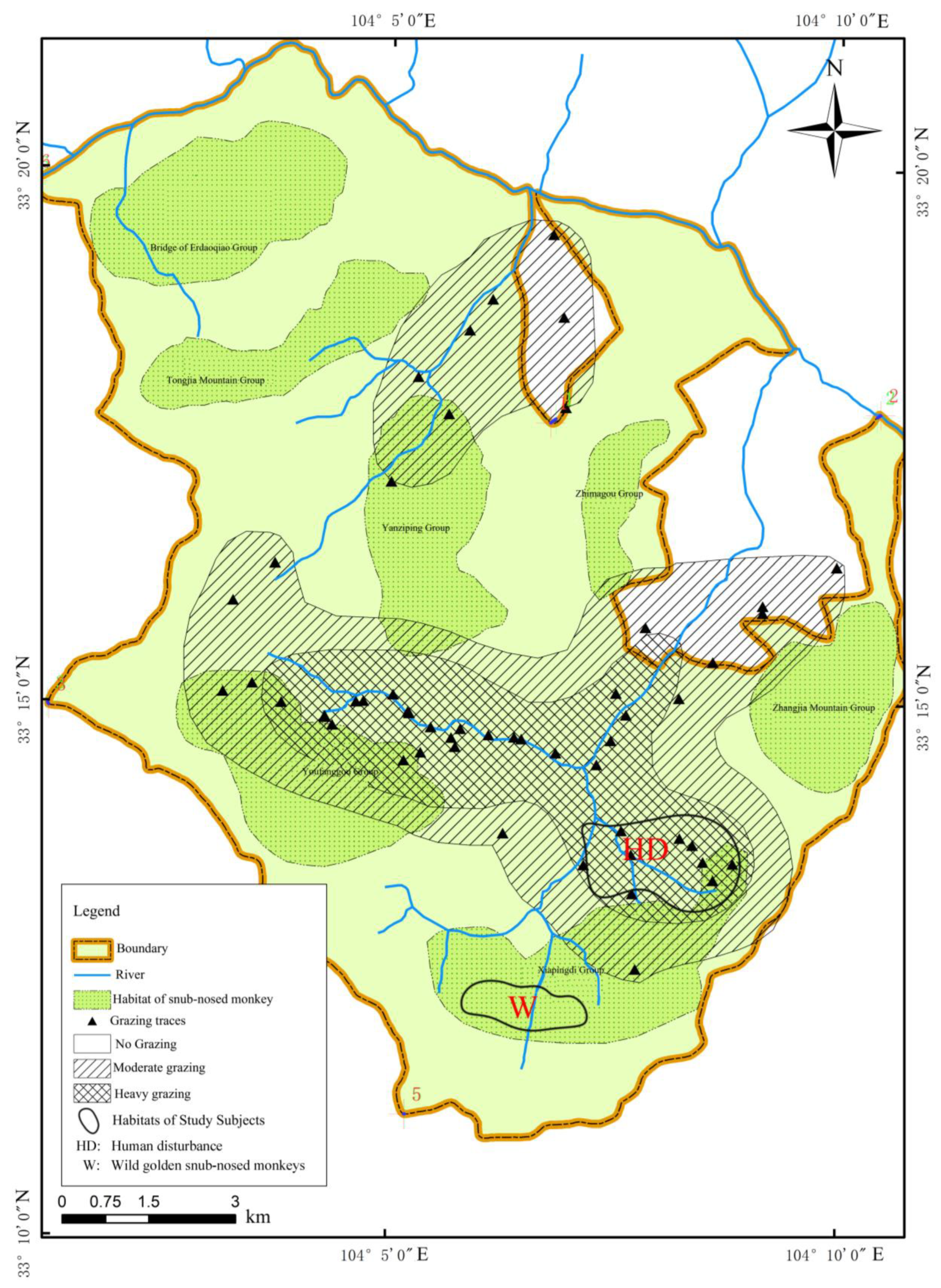
2.1. Study Subjects and Samples Collection
2.2. DNA Extraction
2.3. Analysis of 16S rRNA and Metagenomic Sequencing
2.4. Assembly of Metagenome Assembly Genomes (MAGs)
2.5. Health Risk of PARBs to Golden Snub-Nosed Monkeys
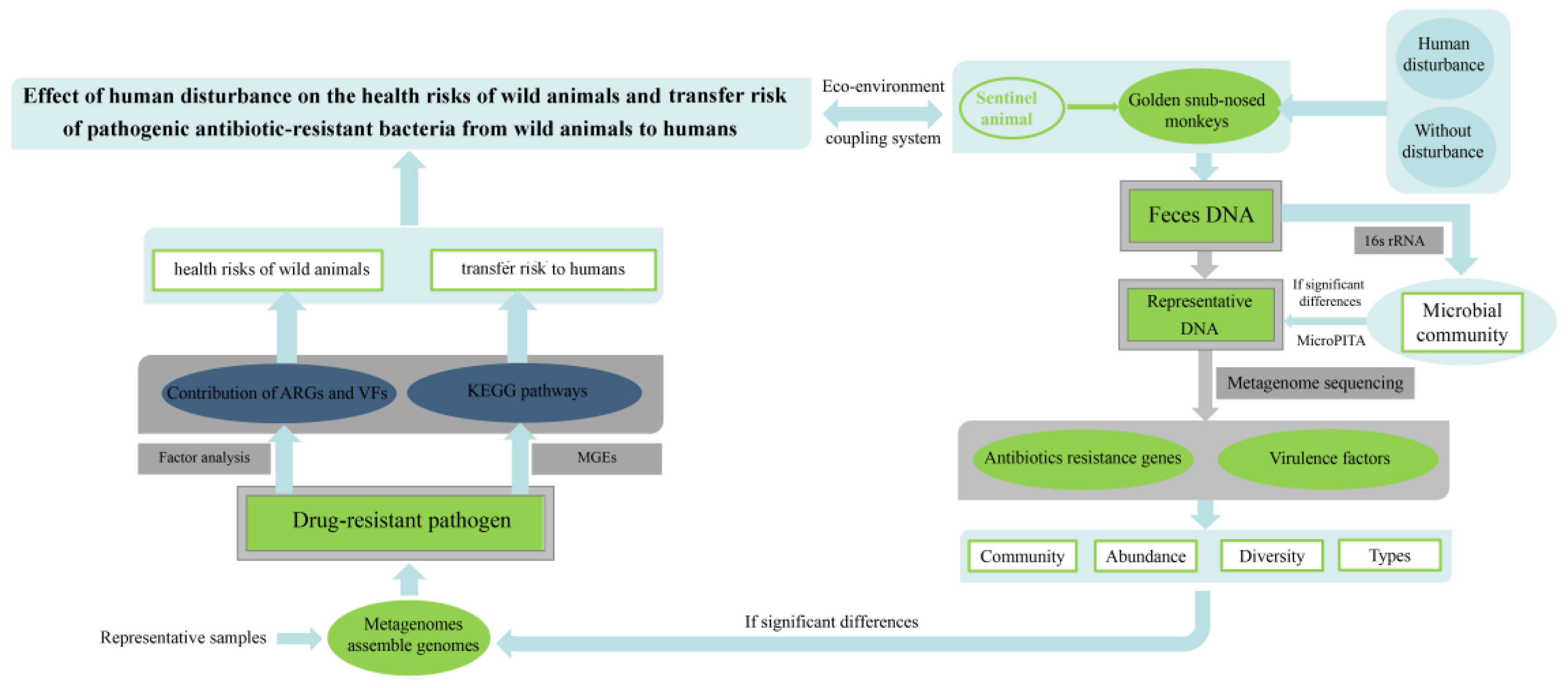
2.6. Research Techniques
3. Results
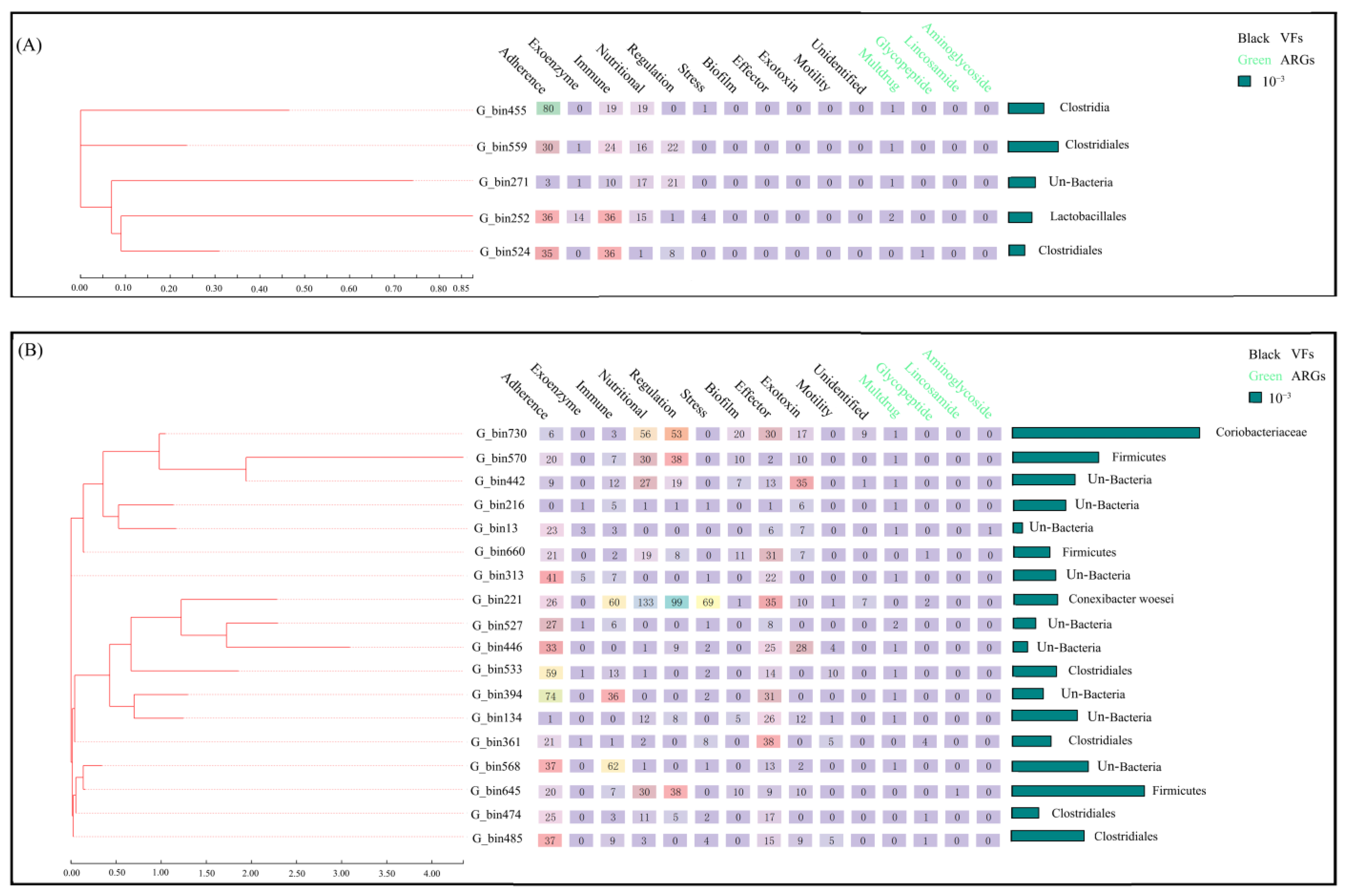
3.1. The Effect of HD on ARGs and VFs in the Gut Microbiota of Golden Snub-Nosed Monkeys
3.2. The Effect of HD on PARBs in the Gut Microbiota of Golden Snub-Nosed Monkeys
3.3. The Effect of HD on Health Risks to Golden Snub-Nosed Monkeys
3.4. The Effect of HD on the Transfer Risk of PARBs from Golden Snub-Nosed Monkeys to Humans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Buta, M.; Korzeniewska, E.; Harnisz, M.; Hubeny, J.; Zieliński, W.; Rolbiecki, D.; Bajkacz, S.; Felis, E.; Kokoszka, K. Microbial and chemical pollutants on the manure-crops pathway in the perspective of “One Health” holistic approach. Sci. Total Environ. 2021, 785, 147411. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.; Song, Q. Legislation advancement of one health in China in the context of the COVID-19 pandemic: From the perspective of the wild animal conservation law. One Health 2021, 12, 100195. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Swaisgood, R.R.; Pilfold, N.W.; Owen, M.A.; Dai, Q.; Wei, F.; Han, H.; Yang, Z.S.; Yang, X.Y.; Gu, X.D.; et al. Assessing the Effectiveness of China’s Panda Protection System. Curr. Biol. 2020, 30, 1280–1286.e2. [Google Scholar] [CrossRef] [PubMed]
- State Forestry Administration. The Fourth National Panda Survey Report; State Forestry Administration: Beijing, China, 2016.
- Ewbank, A.C.; Esperón, F.; Sacristán, C.; Sacristán, I.; Cato-Dias, J.L. Seabirds as anthropization indicators in two different tropical biotopes: A one health approach to the issue of antimicrobial resistance genes pollution in oceanic islands. Sci. Total Environ. 2020, 754, 142141. [Google Scholar] [CrossRef] [PubMed]
- Campbell, T.P.; Sun, X.; Patel, V.H.; Sanz, C.; Morgan, D.; Dantas, G. The microbiome and resistome of chimpanzees, gorillas, and humans across host lifestyle and geography. ISME J. 2020, 14, 1584–1599. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.F. Research progress in conservation biology of endangered mammals in China. Acta Theriol. Sin. 2016, 36, 255–269. [Google Scholar]
- Zhu, D.; Lu, L.; Zhang, Z.J.; Qi, D.W.; Zhang, M.C.; O’Connor, P.; Wei, F.W.; Zhu, Y.G. Insights into the roles of fungi and protist in the giant panda gut microbiome and antibiotic resistome. Environ. Int. 2021, 155, 106703. [Google Scholar] [CrossRef]
- Chen, J.W.; Li, X.J.; Li, L.; Zhang, T.; Zhang, Q.; Wu, F.M.; Wang, D.Y.; Hu, H.Z.; Tian, C.L.; Liao, D.S.; et al. Coagulation factors VII, IX and X are effective antibacterial proteins against drug-resistant Gram-negative bacteria. Cell Res. 2019, 29, 711–724. [Google Scholar] [CrossRef]
- Zhang, A.N.; Gaston, J.M.; Dai, C.L.; Zhao, S.; Poyet, M.; Groussin, M.; Yin, X.; Li, L.-G.; van Loosdrecht, M.C.M.; Topp, E.; et al. An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat. Commun. 2021, 12, 4765. [Google Scholar] [CrossRef]
- Kim, M.; Park, J.; Kang, M.; Yang, J.; Park, W. Gain and loss of antibiotic resistant genes in multidrug resistant bacteria: One health perspective. J. Microbiol. 2021, 59, 535–545. [Google Scholar] [CrossRef]
- Gortazar, C.; Diez-Delgado, I.; Barasona, J.A.; Vicente, J.; De La Fuente, J.; Boadella, M. The wild side of disease control at the wildlife-livestock-human interface: A review. Front. Vet. Sci. 2015, 1, 27. [Google Scholar] [CrossRef] [PubMed]
- Basu, N.; Scheuhammer, A.M.; Bursian, S.J.; Elliott, J.; Rouvinen-Watt, K.; Chan, H.M. Mink as a sentinel species in environmental health. Environ. Res. 2007, 103, 130–144. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.; Fonseca, C.; Mendo, S.; Caetano, T. A closer look on the variety and abundance of the faecal resistome of wild boar. Environ. Pollut. 2022, 292, 118406. [Google Scholar] [CrossRef] [PubMed]
- Chong, C.W.; Alkatheeri, A.H.S.; Ali, N.; Tay, Z.H.; Lee, Y.L.; Paramasivam, S.J.; Jeevaratnam, K.; Low, W.Y.; Lim, S.H.E. Association of antimicrobial resistance and gut microbiota composition in human and non-human primates at an urban ecotourism site. Gut Pathog. 2020, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.Z.; Lu, T.; Huang, C.L.; Wang, J.; Li, D.Y. Grazing disturbance increased the mobility, pathogenicity and host microbial species of antibiotic resistance genes, and multidrug resistance genes posed the highest risk in the habitats of wild animals. Front. Environ. Sci. Front. 2023, 11, 1109298. [Google Scholar] [CrossRef]
- Peng, J.; Zhao, M.; Guo, X.; Pan, Y.; Liu, Y. Spatial-temporal dynamics and associated driving forces of urban ecological land: A case study in shenzhen city, China. Habitat Int. 2017, 60, 81–90. [Google Scholar] [CrossRef]
- Yu, L.; Huang, Y.H.; Zhang, Z.G.; Zhu, X.C.; Li, Y.T.; Li, N.; Liu, C.B.; Xu, Z.R.; Yao, X.Q.; Hu, J.Y.; et al. Technical Specification for Obtaining Intestinal Microbiome Data of Yunnan Snub-Nosed Monkeys. DB53/T 1057-2021. 2021. Available online: http://amr.yn.gov.cn (accessed on 14 October 2021).
- Guo, S.T.; Hou, R.; Garber, P.A.; Raubenheimer, D.; Righini, N.; Ji, W.; Jay, O.; He, S.; Wu, F.; Li, F.; et al. Nutrient-specific compensation for seasonal cold stress in a free-ranging temperate colobine monkey. Funct. Ecol. 2018, 32, 2170–2180. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Yourstone, S.; Mieczkowski, P.; Jones, C.D.; Dangl, J.L. Practical innovations for high-throughput amplicon sequencing. Nat. Methods 2013, 10, 999. [Google Scholar] [CrossRef]
- Zhu, D.; An, X.L.; Chen, Q.L.; Yang, X.R.; Christie, P.; Ke, X.; Wu, L.H.; Zhu, Y.G. Antibiotics Disturb the Microbiome and Increase the Incidence of Resistance Genes in the Gut of a Common Soil Collembolan. Environ. Sci. Technol. 2018, 52, 3081–3090. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic, Note: A Place for DNA-DNA Reassociation and 16S rRNA Sequence Analysis in the Present Species Definition in Bacteriology. Int. J. Syst. Bacteriol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Vatanen, T.; Kostic, A.D.; d’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Timothy, D.A.; Hämäläinen, A.; et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Johnston, E.R.; Barberán, A.; Ren, Y.; Lü, X.; Han, X. Decreased plant productivity resulting from plant group removal experiment constrains soil microbial functional diversity. Glob. Chang. Biol. 2017, 23, 4318–4332. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Alexandre, L.; Mark, B. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010, 38, e132. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.P.; Xia, Y.; Li, B.; Yang, Y.; Li, L.G.; Tiedje, J.M.; Zhang, T. Metagenomic assembly reveals hosts of antibiotic resistance genes and the shared resistome in pig, chicken, and human Feces. Environ. Sci. Technol. 2016, 50, 420–427. [Google Scholar] [CrossRef]
- Xiong, W.G.; Wang, Y.L.; Sun, Y.X.; Ma, L.P.; Zeng, Q.L.; Jiang, X.T.; Li, A.; Zeng, Z.; Zhang, T. Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome 2018, 6, 34. [Google Scholar] [CrossRef]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. Peerj 2015, 3, e1165. [Google Scholar] [CrossRef]
- Uritskiy, G.V.; DiRuggiero, J.; Taylor, J. MetaWRAP-a fexible pipeline for genome-resolved metagenomic data analysis. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Jin, L.; Xie, J.W.; Liu, H.; Zhao, J.; Ye, D.; Li, X.D. Inhalable antibiotic resistomes emitted from hospitals: Metagenomic insights into bacterial hosts, clinical relevance, and environmental risks. Microbiome 2022, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.H.; Wang, Q.J.; Liang, J.H.; Liaokai, L.L.; Qu, J.H. A Method for Evaluating Water Health Risk Based on Antibiotics Resistance Gene and Virulence Factors. National Invention Patent 20201068, 17 November 2020. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Wagner, H. Vegan: Community Ecology Package. R Packag 2011, 2, 5–6. [Google Scholar]
- Wang, Y.T.; Yang, X.Y.; Zhang, M.Y.; Pan, H.J. Comparative Analysis of Gut Microbiota between Wild and Captive Golden Snub-Nosed Monkeys. Animals 2023, 13, 1625. [Google Scholar] [CrossRef] [PubMed]
- Costea, P.I.; Hildebrand, F.; Arumugam, M.; Bäckhed, F.; Blaser, M.J.; Bushman, F.D.; de Vos, W.M.; Ehrlich, S.D.; Fraser, C.M.; Hattori, M.; et al. Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 2017, 3, 8–16. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Zhang, Q.; Wang, T.Z.; Xu, N.H.; Lu, T.; Hong, W.J.; Penuelas, J.; Gillings, M.; Wang, M.X.; Gao, W.W.; et al. Assessment of global health risk of antibiotic resistance genes. Nat. Commun. 2022, 13, 1553. [Google Scholar] [CrossRef]
- Xia, W.C.; Li, G.Q.; Wang, D.L.; Chen, H.; Zhu, L.F.; Li, D.Y. Functional convergence of Yunnan snub-nosed monkey and bamboo-eating panda gut microbiomes revealing driving by dietary flexibility on mammal gut microbiome. Comput. Struct. Biotechnol. J. 2022, 20, 685–699. [Google Scholar] [CrossRef]
- Ju, F.; Beck, K.; Yin, X.; Maccagnan, A.; McArdell, C.S.; Singer, H.P.; Johnson, D.R.; Zhang, T.; Bürgmann, H. Wastewater treatment plant resistomes are shaped by bacterial composition, genetic exchange, and upregulated expression in the effluent microbiomes. ISME J 2019, 13, 346–360. [Google Scholar] [CrossRef]
- Yang, Y.Y.; Liu, G.H.; Ye, C.; Liu, W.Z. Bacterial community and climate change implication affected the diversity and abundance of antibiotic resistance genes in wetlands on the Qinghai-Tibetan Plateau. J. Hazard. Mater. 2019, 361, 283–293. [Google Scholar] [CrossRef]
- Zou, S.Z.; Li, D.Y.; Yuan, T.T.; Zhang, Y.L.; Tang, Y.; Li, L. A Method for Assessing the Effects of Human Disturbance on Wildlife Health Risks Based on a Golden Snub-Noseds Monkeys Biological Model. CN202111521395.6. 2022. Available online: https://d.wanfangdata.com.cn/patent/CN202111521395.6 (accessed on 11 March 2022).
- Zou, S.Z.; Luo, Y.; Cheng, M.; Wang, F.; Li, D.Y.; Kang, D.; Tang, Y. Characteristics of tetracycline antibiotic resistance genes in intestinal microorganisms and intestinal environment of Yunnan snub-nosed monkeys. Acta Theriol. Sin. 2023, 43, 304–314. [Google Scholar]
- Tan, G.; Hu, M.; Li, X.; Pan, Z.; Li, M.; Li, L.; Zheng, Z.; Yang, M. Metagenomics reveals the diversity and taxonomy of antibiotic resistance genes in sufu bacterial communities. Food Control 2021, 121, 107641. [Google Scholar] [CrossRef]
- Zhao, R.; Yu, K.; Zhang, J.; Zhang, G.; Huang, J.; Ma, L.; Deng, C.; Li, X.; Li, B. Deciphering the mobility and bacterial hosts of antibiotic resistance genes under antibiotic selection pressure by metagenomic assembly and binning approaches. Water Res. 2020, 186, 116318. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhou, Y.; Fu, H.; Xiong, X.; Fang, S.; Jiang, H.; Wu, J.; Yang, H.; Gao, J.; Huang, L. Expanded catalog of microbial genes and metagenome-assembled genomes from the pig gut microbiome. Nat. Commun. 2021, 12, 1106. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Qu, Q.; Wang, M.; Huang, M.; Zhou, W.; Wei, F. Global landscape of gut microbiome diversity and antibiotic resistomes across vertebrates. Sci. Total Environ. 2022, 838 (Pt 2), 156178. [Google Scholar] [CrossRef] [PubMed]
- Grilo, M.; Santos, C.S.; Robalo, J.; Oliveira, M. The potential of aeromonas spp. from wildlife as antimicrobial resistance indicators in aquatic environments. Ecol. Indic. 2020, 115, 106396.1–106396.7. [Google Scholar] [CrossRef]
- Densmore, C.L.; Green, D.E. Diseases of amphibians. ILAR J 2007, 48, 235–254. [Google Scholar] [CrossRef]
- Dunn, S.J.; Connor, C.; McNally, A. The evolution and transmission of multi-drug resistant Escherichia coli and Klebsiella pneumoniae: The complexity of clones and plasmids. Curr. Opin. Microbiol. 2019, 51, 51–56. [Google Scholar] [CrossRef]
- Lu, L.; He, Y.; Peng, C.; Wen, X.; Ye, Y.; Ren, D.; Tang, Y.; Zhu, D. Dispersal of antibiotic resistance genes in an agricultural influenced multi-branch river network. Sci. Total Environ. 2022, 830, 154739. [Google Scholar] [CrossRef]
- Liang, J.S.; Mao, G.N.; Yin, X.L.; Ma, L.P.; Liu, L.; Bai, Y.H.; Zhang, T.; Qu, J.H. Identification and quantification of bacterial genomes carrying antibiotic resistance genes and virulence factor genes for aquatic microbiological risk assessment. Water Res. 2020, 168, 115160. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.; Park, M.; Kong, H.; Segre, J. Temporal stability of the human skin microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef]
- Zou, Y.; Xue, W.; Luo, G.; Deng, Z.; Qin, P.; Guo, R.; Sun, H.; Xia, Y.; Liang, S.; Dai, Y.; et al. 1520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat. Biotechnol. 2019, 37, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q.; Cui, L.Y.; Du, X.P.; Hu, Z.F.; Bie, J.; Xiao, J.-H.; Wang, H.-B.; Zhang, J.T. Comparison of gut microbiota diversity and predicted functions between healthy and diseased captive rana dybowskii. Front. Microbiol. 2020, 11, 2096. [Google Scholar] [CrossRef] [PubMed]


| Contig | Antibiotic Resistance Genes (ARGs) | ARG Class | Virulence Factors (VFs) | Mobile Genetic Element Genes (MGEs) | Contig Numbers in Wild Individuals (W Group) | Contig Numbers in Human Disturbance Individuals (HD Group) |
|---|---|---|---|---|---|---|
| HD50_k97_208870_1 | acrB | Multidrug | AcrAB | Plasmid | 6 | 0 |
| HD7_k97_107630_1 | ceoB | Multidrug | AdeFGH | Plasmid | 54 | 2 |
| HD7_k97_185188_1 | adeF | Multidrug | AdeFGH | Plasmid | 2 | 0 |
| HD7_k97_208861_1 | ceoB | Multidrug | AdeFGH | Plasmid | 6 | 0 |
| HD7_k97_226438_1 | ceoB | Multidrug | AdeFGH | Plasmid | 10 | 2 |
| HD7_k97_309577_1 | ceoB | Multidrug | AdeFGH | Plasmid | 92 | 0 |
| HD7_k97_315010_1 | adeF | Multidrug | AdeFGH | Plasmid | 18 | 0 |
| HD7_k97_334322_1 | adeF | Multidrug | AdeFGH | Plasmid | 18 | 80 |
| HD7_k97_375936_1 | adeF | Multidrug | AdeFGH | Plasmid | 6 | 0 |
| HD7_k97_401148_1 | adeF | Multidrug | AdeFGH | Plasmid | 24 | 0 |
| W34_k97_1150608_1 # | MexB | Multidrug | AcrAB | Plasmid | 0 | 12 |
| W34_k97_132510_1 | adeF | Multidrug | AdeFGH | Plasmid | 0 | 12 |
| W34_k97_327771_1 | acrB | Multidrug | AcrAB | Plasmid | 4 | 58 |
| W34_k97_36958_1 | acrB | Multidrug | AcrAB | Plasmid | 0 | 4 |
| W34_k97_551122_1 # | adeF | Multidrug | AdeFGH | Plasmid | 0 | 6 |
| W34_k97_58465_1 | ceoB | Multidrug | AdeFGH | Plasmid | 0 | 12 |
| W34_k97_590319_1 | adeF | Multidrug | AdeFGH | Plasmid | 2 | 12 |
| W34_k97_713320_1 | adeF | Multidrug | AdeFGH | Plasmid | 0 | 24 |
| W34_k97_727054_1 | ceoB | Multidrug | AdeFGH | Plasmid | 0 | 20 |
| W34_k97_939152_1 # | adeF | Multidrug | AdeFGH | Plasmid | 0 | 6 |
| W42_k97_49042_1 # | ugd | Peptide | Capsule | Plasmid | 0 | 6 |
| W42_k97_62472_1 | smeE | Multidrug | AcrAB | Plasmid | 2 | 24 |
| W44_k97_574071_1 | acrB | Multidrug | AcrAB | Plasmid | 2 | 18 |
| W7_k97_500225_1 | ugd | Peptide | Capsule | Plasmid | 0 | 16 |
| Mobility Rate (%) | 58.33 | 78.33 |
| Groups | F1 | F2 | F3 | F4 | F5 | F6 | TF | HR (10−3) | THR (10−3) | |
|---|---|---|---|---|---|---|---|---|---|---|
| W | G_bin455 | −0.267 | −0.667 | 0.026 | −0.180 | −1.940 | 0.078 | −0.3746 | −11.10 | −28.5 |
| G_bin559 | 0.013 | −0.793 | −0.480 | −0.082 | −0.370 | 0.081 | −0.2138 | −8.90 | ||
| G_bin271 | −0.127 | −0.657 | −0.602 | −0.029 | 0.727 | 0.008 | −0.1319 | −3.00 | ||
| G_bin252 | 0.247 | −1.437 | −2.218 | 2.150 | 0.862 | 1.010 | −0.0489 | −0.960 | ||
| G_bin524 | −0.120 | −0.135 | −0.900 | −0.989 | −0.577 | −0.125 | −0.3362 | −4.45 | ||
| HD | G_bin730 | 1.333 | −0.191 | 2.278 | 0.829 | 1.35 | 0.811 | 0.8834 | 138.6 | 125.8 |
| G_bin570 | 0.120 | −0.787 | 0.652 | −0.148 | 0.417 | 0.257 | 0.0455 | 3.280 | ||
| G_bin442 | 0.081 | −0.299 | 1.700 | 0.455 | 0.491 | −0.085 | 0.288 | 15.05 | ||
| G_bin216 | −0.500 | −0.448 | −0.346 | 0.140 | 0.880 | −0.079 | −0.1362 | −5.96 | ||
| G_bin13 | −0.331 | −0.569 | −0.002 | 0.290 | 0.394 | −4.322 | −0.3845 | −3.02 | ||
| G_bin660 | −0.293 | 0.404 | 0.756 | −0.547 | 0.559 | 0.336 | 0.1038 | 3.14 | ||
| G_bin313 | −0.417 | −0.068 | −0.636 | 0.661 | 0.171 | 0.432 | −0.0822 | −2.90 | ||
| G_bin221 | 4.143 | 0.933 | −0.579 | −0.197 | −0.626 | −0.462 | 0.9991 | 36.7 | ||
| G_bin527 | −0.504 | −0.584 | −0.213 | 0.877 | 0.071 | 0.276 | −0.1224 | −2.29 | ||
| G_bin446 | −0.620 | 0.856 | 1.535 | 0.672 | −0.399 | −0.085 | 0.1856 | 2.25 | ||
| G_bin533 | −0.850 | 1.192 | 0.399 | 0.578 | −1.56 | 0.485 | −0.0619 | −2.27 | ||
| G_bin394 | −0.171 | 0.050 | 0.221 | 0.150 | −2.04 | 0.321 | −0.1812 | −4.67 | ||
| G_bin134 | −0.413 | 0.301 | 0.685 | 0.355 | 0.947 | 0.191 | 0.1735 | 9.48 | ||
| G_bin316 | −0.443 | 3.035 | −1.477 | −0.347 | 1.39 | 0.181 | 0.2539 | 8.21 | ||
| G_bin568 | 0.106 | −0.620 | −0.312 | 0.015 | −1.464 | 0.184 | −0.2394 | −15.24 | ||
| G_bin645 | −0.031 | −1.390 | 0.078 | −3.46 | 0.729 | 0.702 | −0.4365 | −48.4 | ||
| G_bin474 | −0.358 | 0.598 | −0.613 | −0.757 | 0.397 | −0.118 | −0.1294 | −2.91 | ||
| G_bin485 | −0.598 | 1.277 | 0.047 | −0.436 | −0.399 | −0.074 | −0.0536 | −3.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, S.; Yuan, T.; Lu, T.; Yan, J.; Kang, D.; Li, D. Human Disturbance Increases Health Risks to Golden Snub-Nosed Monkeys and the Transfer Risk of Pathogenic Antibiotic-Resistant Bacteria from Golden Snub-Nosed Monkeys to Humans. Animals 2023, 13, 3083. https://doi.org/10.3390/ani13193083
Zou S, Yuan T, Lu T, Yan J, Kang D, Li D. Human Disturbance Increases Health Risks to Golden Snub-Nosed Monkeys and the Transfer Risk of Pathogenic Antibiotic-Resistant Bacteria from Golden Snub-Nosed Monkeys to Humans. Animals. 2023; 13(19):3083. https://doi.org/10.3390/ani13193083
Chicago/Turabian StyleZou, Shuzhen, Tingting Yuan, Tan Lu, Jiayu Yan, Di Kang, and Dayong Li. 2023. "Human Disturbance Increases Health Risks to Golden Snub-Nosed Monkeys and the Transfer Risk of Pathogenic Antibiotic-Resistant Bacteria from Golden Snub-Nosed Monkeys to Humans" Animals 13, no. 19: 3083. https://doi.org/10.3390/ani13193083




