Use of Danish National Somatic Cell Count Data to Assess the Need for Dry-Off Treatment in Holstein Dairy Cattle
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
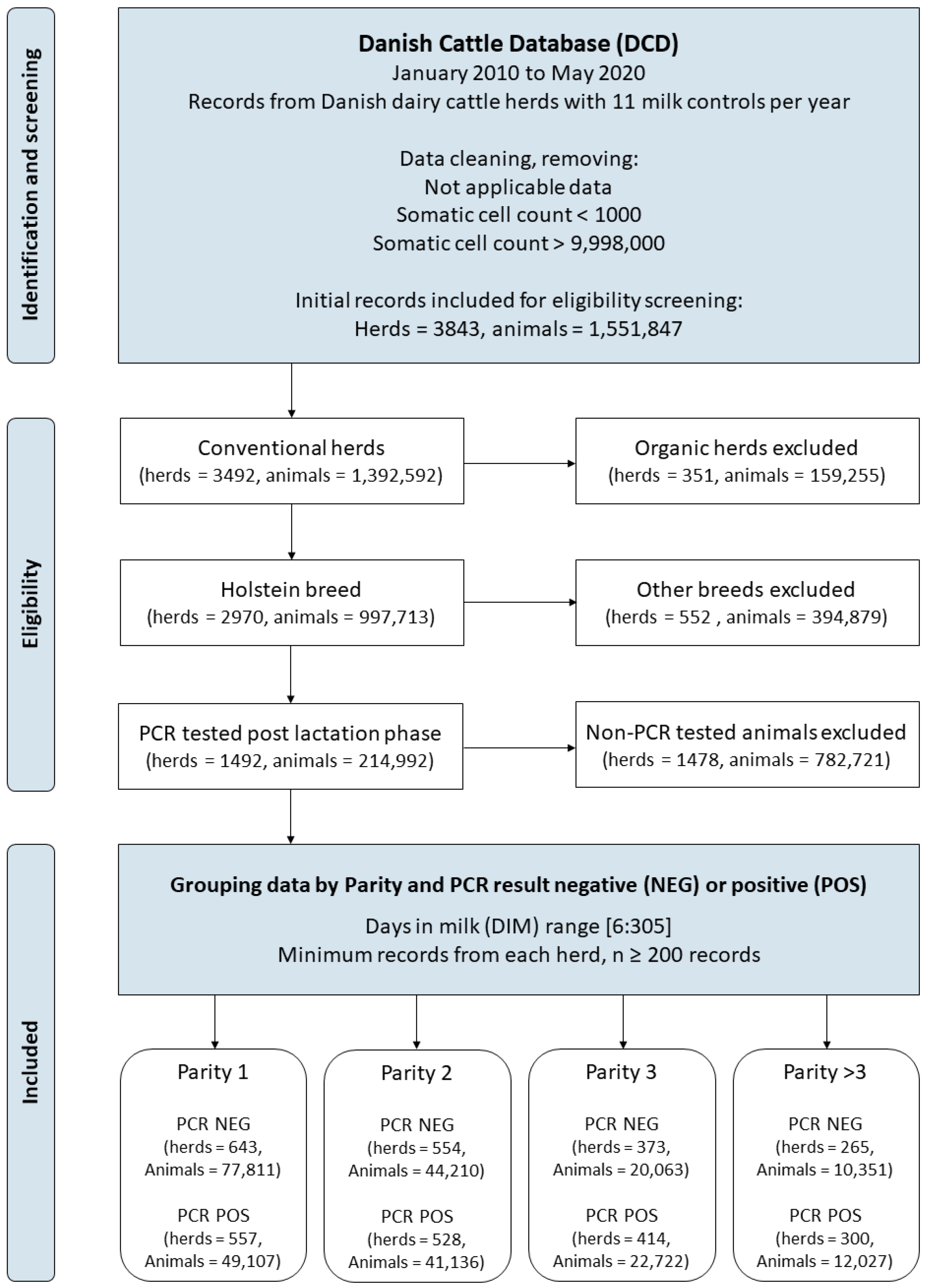
2.1. Data
2.2. Descriptive Analysis
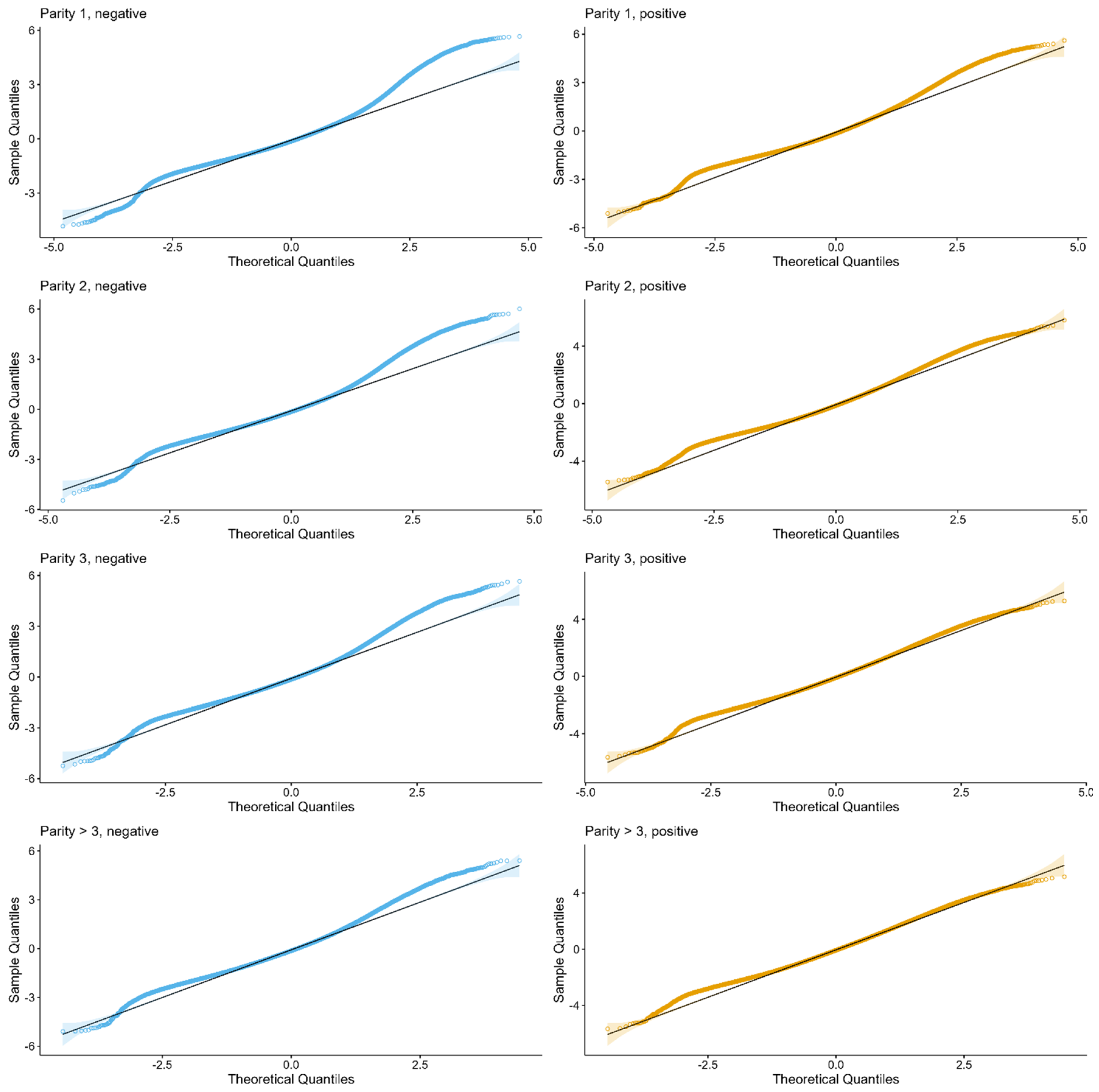
2.3. Somatic Cell Count
3. Results
3.1. Descriptive Analysis
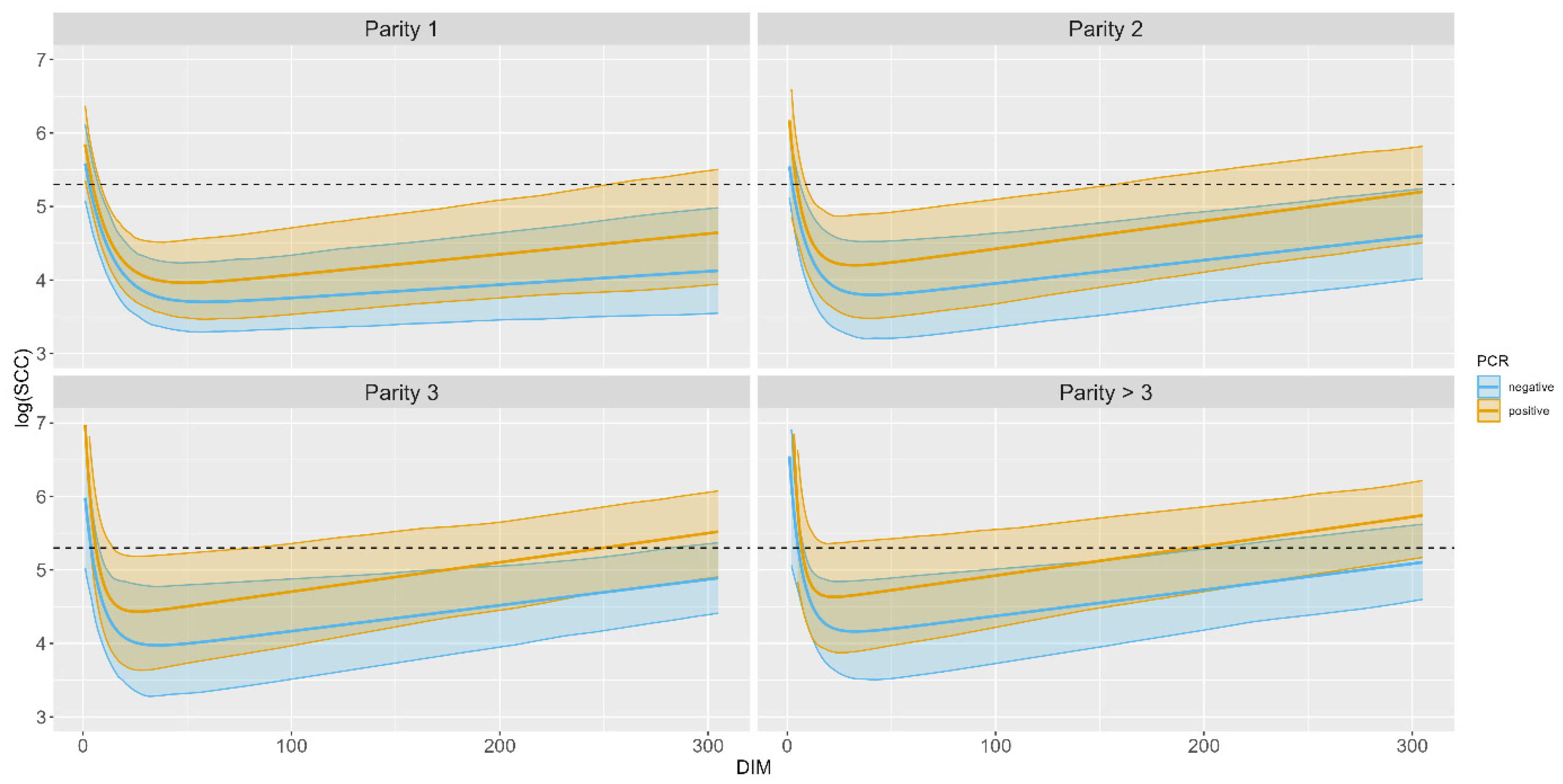
3.2. Somatic Cell Count Modelling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. WHO—Antimicrobial Resistance. 2018. Available online: http://www.who.int/en/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 6 June 2018).
- European Commission. Animal Health Law 2016; European Commission: Brussels, Belgium, 2016. [Google Scholar]
- Kuipers, A.; Koops, W.J.; Wemmenhove, H. Antibiotic use in dairy herds in the Netherlands from 2005 to 2012. J. Dairy Sci. 2016, 99, 1632–1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DANMAP. DANMAP 2019—Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food And Humans in Denmark; Statens Serum Institute: Copenhagen, Denmark, 2019; Available online: https://www.danmap.org/reports/2019 (accessed on 27 July 2023).
- Halasa, T.; Huijps, K.; Østerås, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef] [PubMed]
- van den Borne, B.; Halasa, T.; van Schaik, G.; Hogeveen, H.; Nielen, M. Bioeconomic modeling of lactational antimicrobial treatment of new bovine subclinical intramammary infections caused by contagious pathogens. J. Dairy Sci. 2010, 93, 4034–4044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pantoja, J.C.F.; Hulland, C.; Ruegg, P.L. Somatic cell count status across the dry period as a risk factor for the development of clinical mastitis in the subsequent lactation. J. Dairy Sci. 2009, 92, 139–148. [Google Scholar] [CrossRef] [Green Version]
- Cameron, M.; Keefe, G.P.; Roy, J.P.; Stryhn, H.; Dohoo, I.R.; McKenna, S.L. Evaluation of selective dry cow treatment following on-farm culture: Milk yield and somatic cell count in the subsequent lactation. J. Dairy Sci. 2015, 98, 2427–2436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SEGES. SEGES Antibiotic Usage; Aarhus Universitet: Aarhus, Denmark, 2015; Available online: https://www.landbrugsinfo.dk/-/media/landbrugsinfo/public/0/c/b/kv_17_retningslinjer-for-brug-af-antibiotika.pdf (accessed on 27 July 2023).
- Niemi, R.E.; Hovinen, M.; Rajala-Schultz, P.J. Selective dry cow therapy effect on milk yield and somatic cell count: A retrospective cohort study. J. Dairy Sci. 2022, 105, 1387–1401. [Google Scholar] [CrossRef]
- Taponen, S.; Liski, E.; Heikkilä, A.M.; Pyörälä, S. Factors associated with intramammary infection in dairy cows caused by coagulase-negative staphylococci, Staphylococcus aureus, Streptococcus uberis, Streptococcus dysgalactiae, Corynebacterium bovis, or Escherichia coli. J. Dairy Sci. 2017, 100, 493–503. [Google Scholar] [CrossRef] [Green Version]
- Tenhagen, B.A.; Köster, G.; Wallmann, J.; Heuwieser, W. Prevalence of Mastitis Pathogens and Their Resistance against Antimicrobial Agents in Dairy Cows in Brandenburg, Germany. J. Dairy Sci. 2006, 89, 2542–2551. [Google Scholar] [CrossRef] [Green Version]
- Rysanek, D.; Zouharova, M.; Babak, V. Monitoring major mastitis pathogens at the population level based on examination of bulk tank milk samples. J. Dairy Res. 2009, 76, 117–123. [Google Scholar] [CrossRef]
- Cederlöf, S.E.; Toft, N.; Aalbaek, B.; Klaas, I.C. Latent class analysis of the diagnostic characteristics of PCR and conventional bacteriological culture in diagnosing intramammary infections caused by Staphylococcus aureus in dairy cows at dry off. Acta Vet. Scand. 2012, 54, 65. [Google Scholar] [CrossRef] [Green Version]
- Mahmmod, Y.S.; Mweu, M.M.; Nielsen, S.S.; Katholm, J.; Klaas, I.C. Effect of carryover and presampling procedures on the results of real-time PCR used for diagnosis of bovine intramammary infections with Streptococcus agalactiae at routine milk recordings. Prev. Vet. Med. 2014, 113, 512–521. [Google Scholar] [CrossRef]
- Mahmmod, Y.S.; Klaas, I.C.; Nielsen, S.S.; Katholm, J.; Toft, N. Effect of presampling procedures on real-time PCR used for diagnosis of intramammary infections with Staphylococcus aureus in dairy cows at routine milk recordings. J. Dairy Sci. 2013, 96, 2226–2233. [Google Scholar] [CrossRef] [Green Version]
- Schukken, Y.H.; Wilson, D.J.; Welcome, F.; Garrison-Tikofsky, L.; Gonzalez, R.N. Monitoring udder health and milk quality using somatic cell counts. Vet. Res. 2003, 34, 579–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkeby, C.; Toft, N.; Schwarz, D.; Farre, M.; Nielsen, S.; Zervens, L.; Hechinger, S.; Halasa, T. Differential somatic cell count as an additional indicator for intramammary infections in dairy cows. J. Dairy Sci. 2020, 103, 1759–1775. [Google Scholar] [CrossRef] [Green Version]
- Madouasse, A.; Huxley, J.N.; Browne, W.J.; Bradley, A.J.; Green, M.J. Somatic cell count dynamics in a large sample of dairy herds in England and Wales. Prev. Vet. Med. 2010, 96, 56–64. [Google Scholar] [CrossRef]
- Bradley, A.; Green, M. Use and interpretation of somatic cell count data in dairy cows. Practice 2005, 27, 310–315. [Google Scholar] [CrossRef]
- Dohoo, I.R.; Leslie, K.E. Evaluation of changes in somatic cell counts as indicators of new intramammary infections. Prev. Vet. Med. 1991, 10, 225–237. [Google Scholar] [CrossRef]
- Kirkeby, C.; Schwarz, D.; Denwood, M.; Farre, M.; Nielsen, S.S.; Gussmann, M.; Toft, N.; Halasa, T. Dynamics of somatic cell count (SCC) and differential SCC during and following intramammary infections. J. Dairy Sci. 2021, 104, 3427–3438. [Google Scholar] [CrossRef] [PubMed]
- Zecconi, A.; Dell’orco, F.; Vairani, D.; Rizzi, N.; Cipolla, M.; Zanini, L. Differential somatic cell count as a marker for changes of milk composition in cows with very low somatic cell count. Animals 2020, 10, 604. [Google Scholar] [CrossRef] [Green Version]
- Riekerink, R.G.M.O.; Barkema, H.W.; Veenstra, W.; Berg, F.E.; Stryhn, H.; Zadoks, R.N. Somatic Cell Count during and between Milkings. J. Dairy Sci. 2007, 90, 3733–3741. [Google Scholar] [CrossRef] [Green Version]
- Shook, G.E.; Kirk, R.L.B.; Welcome, F.L.; Schukken, Y.H.; Ruegg, P.L. Relationship between intramammary infection prevalence and somatic cell score in commercial dairy herds. J. Dairy Sci. 2017, 100, 9691–9701. [Google Scholar] [CrossRef] [PubMed]
- Sumon, S.M.M.R.; Parvin, M.S.; Ehsan, M.A.; Islam, M.T. Dynamics of somatic cell count and intramammary infection in lactating dairy cows. J. Adv. Vet. Anim. Res. 2020, 7, 314. [Google Scholar] [CrossRef] [PubMed]
- Græsbøll, K.; Kirkeby, C.; Nielsen, S.S.; Halasa, T.; Toft, N.; Christiansen, L.E. Models to Estimate Lactation Curves of Milk Yield and Somatic Cell Count in Dairy Cows at the Herd Level for the Use in Simulations and Predictive Models. Front. Vet. Sci. 2016, 3, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SEGES. Nye Regler for Gold-Og Yverbehandlinger af Malkekvæg. 2021. Available online: https://www.landbrugsinfo.dk/public/0/a/0/sundhed_velfard_nye_-regler_gold_-yverbehandlinger (accessed on 14 October 2021).
- Wilmink, J.B.M. Comparison of different methods of predicting 305-day milk yield using means calculated from within-herd lactation curves. Livest. Prod. Sci. 1987, 17, 1–17. [Google Scholar] [CrossRef]
- RYK Fonden. PCR—Test for Yverinfektion før Goldbehandling; RYK Fonden: Aarhus, Denmark, 2019; pp. 2–4. [Google Scholar]
- Koskinen, M.T.; Holopainen, J.; Pyörälä, S.; Bredbacka, P.; Pitkälä, A.; Barkema, H.W.; Bexiga, R.; Roberson, J.; Sølverød, L.; Piccinini, R.; et al. Analytical specificity and sensitivity of a real-time polymerase chain reaction assay for identification of bovine mastitis pathogens. J. Dairy Sci. 2009, 92, 952–959. [Google Scholar] [CrossRef]
- SEGES. Fakta om Mælkeproduktion|Kvæg|SEGES. 2021. Available online: https://www.seges.dk/fagomraader/kvaeg/tal-og-fakta-om-kvaegproduktion/maelkeproduktion#Danskemalkekvægsracer (accessed on 29 December 2021).
- L&F. Kvæg. 2017. Available online: https://lf.dk/viden-om/landbrugsproduktion/husdyr/kvag (accessed on 25 May 2021).
- Riekerink, R.G.M.O.; Kelton, D.; Gurjar, A.; Middleton, J. The use of PCR to diagnose the cause of intramammary infections in dairy cattle. 2019. Available online: https://www.nmconline.org/wp-content/uploads/2019/01/PCR-Fact-Sheet-for-NMC-updated-09_01_2019.pdf (accessed on 27 July 2023).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.r-project.org (accessed on 27 July 2023).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; R Core Team. Nlme: Linear and Nonlinear Mixed Effects Models. 2021. Available online: https://cran.r-project.org/web/packages/nlme/index.html (accessed on 27 July 2023).
- Bates, D.M.; Chambers, J.M. Nonlinear Models; Chapter 10; Wadsworth & Brooks/Cole: Pacific Grove, CA, USA, 1991. [Google Scholar]
- Padfield, D.; Matheso, G. Package ‘Nls.multstart; CRAN: Vienna, Austria, 2020; pp. 1–5. [Google Scholar]
- Mooney, S.J.; Westreich, D.J.; El-Sayed, A.M. Epidemiology in the Era of Big Data. Epidemiology 2015, 26, 390. [Google Scholar] [CrossRef]
- Patel, K.; Godden, S.M.; Royster, E.; Crooker, B.A.; Timmerman, J.; Fox, L. Relationships among bedding materials, bedding bacteria counts, udder hygiene, milk quality, and udder health in US dairy herds. J. Dairy Sci. 2019, 102, 10213–10234. [Google Scholar] [CrossRef]
- Santman-Berends, I.M.G.A.; Swinkels, J.M.; Lam, T.J.G.M.; Keurentjes, J.; van Schaik, G. Evaluation of udder health parameters and risk factors for clinical mastitis in Dutch dairy herds in the context of a restricted antimicrobial usage policy. J. Dairy Sci. 2016, 99, 2930–2939. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, X.; Yang, F.; Luo, J.; Wang, X.; Liu, L.; Li, H. Influences of season, parity, lactation, udder area, milk yield, and clinical symptoms on intramammary infection in dairy cows. J. Dairy Sci. 2016, 99, 6484–6493. [Google Scholar] [CrossRef]
- Wood, P.D.P. Algebraic model of the lactation curve in cattle. Nature 1967, 216, 164–165. [Google Scholar] [CrossRef]
- Gussmann, M.; Græsbøll, K.; Toft, N.; Nielsen, S.S.; Farre, M.; Kirkeby, C.; Halasa, T. Determinants of antimicrobial treatment for udder health in Danish dairy cattle herds. J. Dairy Sci. 2018, 101, 505–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Haas, Y.; Veerkamp, R.F.; Barkema, H.W.; Gröhn, Y.T.; Schukken, Y.H. Associations between pathogen-specific cases of clinical mastitis and somatic cell count patterns. J. Dairy Sci. 2004, 87, 95–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Haas, Y.; Barkema, H.W.; Veerkamp, R.F. The effect of pathogen-specific clinical mastitis on the lactation curve for somatic cell count. J. Dairy Sci. 2002, 85, 1314–1323. [Google Scholar] [CrossRef]
- Laevens, H.; Deluyker, H.; Schukken, Y.; De Meulemeester, L.; Vandermeersch, R.; De Muêlenaere, E.; De Kruif, A. Influence of Parity and Stage of Lactation on the Somatic Cell Count in Bacteriologically Negative Dairy Cows. J. Dairy Sci. 1997, 80, 3219–3226. [Google Scholar] [CrossRef]
- Petzer, I.M.; Karzis, J.; Donkin, E.F.; Webb, E.C.; Etter, E.M.C. Validity of somatic cell count as indicator of pathogen-specific intramammary infections. J. South Afr. Vet. Assoc. 2017, 88, 1019–9128. [Google Scholar] [CrossRef] [Green Version]
- Mahmmod, Y.S.; Klaas, I.C.; Enevoldsen, C. DNA carryover in milk samples from routine milk recording used for PCR-based diagnosis of bovine Staphylococcus aureus mastitis. J. Dairy Sci. 2017, 100, 5709–5716. [Google Scholar] [CrossRef] [Green Version]
- Thygesen, L.C.; Ersbøll, A.K. When the entire population is the sample: Strengths and limitations in register-based epidemiology. Eur. J. Epidemiol. 2014, 29, 551–558. [Google Scholar] [CrossRef]

| Parity | PCR | Total SCC Observations | Total Animals | Geometric Mean | Min | Q1 | Median | Q3 | Max |
|---|---|---|---|---|---|---|---|---|---|
| 1 | NEG | 667,336 | 77,811 | 46 | 1 | 22 | 40 | 83 | 9978 |
| POS | 420,129 | 49,107 | 70 | 1 | 28 | 59 | 149 | 9986 | |
| 2 | NEG | 381,379 | 44,210 | 60 | 1 | 26 | 54 | 120 | 9982 |
| POS | 353,818 | 41,136 | 103 | 1 | 37 | 95 | 250 | 9997 | |
| 3 | NEG | 173,335 | 20,063 | 77 | 1 | 32 | 70 | 164 | 9997 |
| POS | 195,845 | 22,722 | 138 | 1 | 51 | 136 | 347 | 9998 | |
| >3 | NEG | 104,673 | 10,351 | 97 | 1 | 38 | 91 | 218 | 9975 |
| POS | 120,395 | 12,027 | 171 | 1 | 62 | 172 | 444 | 9996 |
| Param | Parity | PCR | Mean | Median | SD |
|---|---|---|---|---|---|
| a | 1 | NEG | 3.57 | 3.56 | 0.229 |
| POS | 3.79 | 3.78 | 0.268 | ||
| 2 | NEG | 3.64 | 3.61 | 0.368 | |
| POS | 4.04 | 4.05 | 0.387 | ||
| 3 | NEG | 3.81 | 3.80 | 0.399 | |
| POS | 4.31 | 4.32 | 0.413 | ||
| >3 | NEG | 4.02 | 4.02 | 0.398 | |
| POS | 4.52 | 4.56 | 0.414 | ||
| b | 1 | NEG | 1.82 × 10−3 | 1.70 × 10−3 | 6.98 × 10−4 |
| POS | 2.81 × 10−3 | 2.73 × 10−3 | 7.53 × 10−4 | ||
| 2 | NEG | 3.17 × 10−3 | 3.23 × 10−3 | 6.84 × 10−4 | |
| POS | 3.79 × 10−3 | 3.79 × 10−3 | 8.04 × 10−4 | ||
| 3 | NEG | 3.53 × 10−3 | 3.54 × 10−3 | 7.53 × 10−4 | |
| POS | 3.98 × 10−3 | 3.99 × 10−3 | 9.22 × 10−4 | ||
| >3 | NEG | 3.57 × 10−3 | 3.60 × 10−3 | 7.84 × 10−4 | |
| POS | 3.99 × 10−3 | 4.05 × 10−3 | 9.92 × 10−4 | ||
| c | 1 | NEG | −2.54 | −2.54 | 0.114 |
| POS | −2.43 | −2.42 | 0.214 | ||
| 2 | NEG | −2.25 | −2.25 | 0.137 | |
| POS | −1.93 | −1.93 | 0.310 | ||
| 3 | NEG | −2.03 | −2.01 | 0.293 | |
| POS | −1.62 | −1.61 | 0.148 | ||
| >3 | NEG | −1.85 | −1.85 | 0.303 | |
| POS | −1.26 | −1.25 | 0.331 | ||
| d | 1 | NEG | 2.17 | 2.18 | 0.232 |
| POS | 2.24 | 2.25 | 0.137 | ||
| 2 | NEG | 2.12 | 2.13 | 0.143 | |
| POS | 2.50 | 2.47 | 0.464 | ||
| 3 | NEG | 2.49 | 2.49 | 0.273 | |
| POS | 3.26 | 3.26 | 0.0170 | ||
| >3 | NEG | 3.00 | 3.01 | 0.532 | |
| POS | 5.97 | 6.03 | 1.91 |
| Parity | PCR | Min logSCC | Min SCC | DIM for Min SCC | ΔlogSCC for DIM Range 100–150 |
|---|---|---|---|---|---|
| 1 | NEG | 3.700256 | 40,458 | 58 | 0.0018011 |
| POS | 3.954595 | 52,175 | 48 | 0.0028032 | |
| 2 | NEG | 3.794039 | 44,436 | 40 | 0.0031664 |
| POS | 4.189145 | 66,966 | 31 | 0.0037942 | |
| 3 | NEG | 3.962009 | 52,563 | 34 | 0.0035282 |
| POS | 4.428367 | 83,794 | 26 | 0.0039846 | |
| >3 | NEG | 4.150237 | 63,449 | 31 | 0.0035651 |
| POS | 4.623989 | 101,900 | 21 | 0.0039937 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Henningsen, M.B.; Denwood, M.; Kirkeby, C.T.; Nielsen, S.S. Use of Danish National Somatic Cell Count Data to Assess the Need for Dry-Off Treatment in Holstein Dairy Cattle. Animals 2023, 13, 2523. https://doi.org/10.3390/ani13152523
Henningsen MB, Denwood M, Kirkeby CT, Nielsen SS. Use of Danish National Somatic Cell Count Data to Assess the Need for Dry-Off Treatment in Holstein Dairy Cattle. Animals. 2023; 13(15):2523. https://doi.org/10.3390/ani13152523
Chicago/Turabian StyleHenningsen, Maj Beldring, Matt Denwood, Carsten Thure Kirkeby, and Søren Saxmose Nielsen. 2023. "Use of Danish National Somatic Cell Count Data to Assess the Need for Dry-Off Treatment in Holstein Dairy Cattle" Animals 13, no. 15: 2523. https://doi.org/10.3390/ani13152523





