Simple Summary
Antimicrobial resistance (AMR) is recognised as an urgent global threat, both in human and in veterinary medicine. In recent years, increasing interest has turned to wild animals in order to evaluate their role as additional sources of antimicrobial resistance. Resistant bacteria may be cycling through wildlife and back into the ecosystem, then wildlife populations may act as sentinels for AMR in the environment. In this study, an epidemiological investigation of the spread of tetracycline resistance (tet) genes in wild birds was performed using a culture-independent approach. Positivity for one or more tet genes was found in 114 (45%) of 254 free-living birds tested. In view of the growing anthropogenic pressure, the spread of antimicrobial resistance in wildlife and the implications for resistance control strategies will have to be considered carefully.
Abstract
Wild animals are less likely to be exposed directly to clinical antimicrobial agents than domestic animals or humans, but they can acquire antimicrobial-resistant bacteria through contact with humans, animals, and the environment. In the present study, 254 dead free-living birds belonging to 23 bird species were examined by PCR for the presence of tetracycline resistance (tet) genes. A fragment of the spleen was collected from each bird carcass. A portion of the intestine was also taken from 73 of the 254 carcasses. Extracted DNA was subjected to PCR amplification targeting the tet(L), tet(M), and tet(X) genes. In total, 114 (45%) of the 254 birds sampled belonging to 17 (74%) of the 23 bird species tested were positive for one or more tet genes. The tet(M) gene showed a higher frequency than the other tested genes, both in the spleen and in the intestine samples. These results confirm the potential role of wild birds as reservoirs, dispersers, or bioindicators of antimicrobial resistance in the environment.
1. Introduction
Antimicrobial resistance (AMR) is a complex phenomenon involving three interdependent ecosystems of potential relevance to public health: humans, animals, and the natural environment. Antimicrobial resistance naturally occurs due to the production of antimicrobial molecules by strains of bacteria and fungi in all environments, including soil [1,2]. However, in the last decades, there has been an acceleration caused by the misuse and overuse of antimicrobials in both human and veterinary medicine, as well as in livestock, where antimicrobials have been used as growth supplements [3]. In the veterinary field, most studies on AMR have focused on the animal species most subjected to pharmacological pressure, such as intensively reared cattle, pigs, poultry, and fish, implicated as reservoirs for multidrug-resistant foodborne pathogens. Usually, wild animals are less likely to be exposed directly to clinical antimicrobial agents than domestic animals or humans [4], except occasionally in rehabilitation facilities. However, wildlife is part of the natural environmental compartment and can be influenced by anthropogenic pressures, e.g., by human waste systems or animal husbandry facilities that can be a source of active antimicrobials, antimicrobial-resistant bacteria, and resistance genes [5].
Conventional antimicrobial susceptibility testing methods are based on bacteriological culture and antimicrobial susceptibility testing of the isolated microorganisms. A limitation of culture-dependent methods is the existence of labile or viable but not culturable bacteria, as well microorganisms that require a long period of growth. Recent studies introduced molecular approaches based on amplification of antimicrobial resistance target genes from environmental or biological samples [6,7,8,9,10]. This approach is more expensive than traditional cultivation and does not allow determination of the bacterial sources of resistance genes. However, it is a fast method and avoids possible underestimation of the occurrence of AMR due to a consistent not culturable or labile fraction of microorganisms [11]. Moreover, considering AMR genes as contamination markers, methods which allow searching for these genes rather than for the bacteria carrying them could aid epidemiological efforts to analyse the spread of resistance determinants [12,13,14].
The aim of this study was to investigate by PCR the presence of tetracycline (tet) resistance genes in free-living birds in Italy.
2. Materials and Methods
2.1. Sampling
From 2016 to 2020, 254 free-living birds, belonging to 23 bird species, that died from trauma or predation (Table 1) were collected at a wildlife recovery centre located in Tuscany (Central Italy) and transported to the Department of Veterinary Sciences of Pisa University (Pisa, Italy) for educational activities. In the present study, only intact organs were examined. A fragment of the spleen was collected from each bird carcass and immediately stored at −20 °C. A portion of the intestine was also taken from 73 of the 254 carcasses.

Table 1.
Bird species sampled.
2.2. Molecular Analysis
2.2.1. DNA Extraction
Total DNA was individually extracted from each sample using the QIAamp DNA mini kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. Negative (kit reagents only) controls were included in each set of extraction. The DNA extracts were stored at −20 °C before analysis.
2.2.2. DNA Amplification and Sequencing
DNA samples were investigated to search for three genes involved in the three tetracycline resistance mechanisms: tet(L) (tetracycline efflux pumps), tet(M) (ribosomal protection) and tet(X) (enzymatic inactivation). Each tet gene was amplified in an individual PCR, using primers described by Ng et al. [15] (Table 2).

Table 2.
Primers used for the detection of tetracycline resistance target genes.
Different PCR protocols were carried out: 5 min of initial denaturation at 94 °C followed by 35 cycles at 94 °C for 1 min; 53 °C (tet(M) and tet(X)) or 55 °C (tet(L)) for 1 min; and 72 °C for 1 min. A final step of 10 min at 72 °C completed the reaction. The DNA extracted from Escherichia coli field strains, containing tetracycline resistance plasmids, was used as a positive control. The extraction control and a distilled water negative control were also included.
The PCR products were analysed by gel electrophoresis (2% agarose); the DNA bands were stained with Midori Green Advance (Nippon Genetics Europe GmbH, Düren, Germany) and then visualised using ultraviolet (UV) trans illumination. The amplicons were purified using the High Pure PCR Product Purification Kit (Roche, Mannheim, Germany), and both DNA strands were sequenced (Bio-Fab Research, Rome, Italy). The sequences obtained were compared with the public sequences available using the BLAST server in GenBank (National Center for Biotechnology Information 2022).
3. Results
In total, of the 254 birds sampled, 114 (45%), belonging to 17 (74%) of the 23 bird species tested, were positive for one or more tet genes. With respect to the 254 tested spleens, 82 (32%) were tet(M) positive, 7 (3%) were tet(L) positive, 4 (1.5%) were tet(X) positive and 21 (8%) were positive for both tet(M) and tet(L) (Figure 1; Table 3).
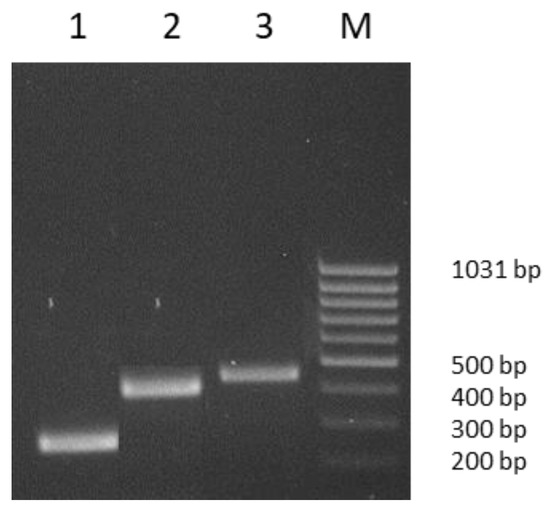
Figure 1.
PCR amplicons. Lane 1, 267 bp tet(L) gene fragment; lane 2, 406 bp tet(M) gene fragment; lane 3, 468 bp tet(X) gene fragment; lane M, MassRuler Low Range DNA Ladder, (Thermo Fisher Scientific, Vilnius, Lithuania).

Table 3.
Tetracycline resistance genes detected in the spleens of wild birds.
With respect to the intestine samples, 18 (25%) of 73 samples collected were positive for one or more tet genes. In particular, 14 (19%) were positive for tet(M), 2 (3%) for tet(L) and 2 (3%) for both tet(M) and tet(L). No positivity for tet(X) was found in intestine samples (Table 4).

Table 4.
Tetracycline resistance genes detected in the intestines of wild birds.
Each tet positive intestine sample was from a carcass whose spleen was positive for one or more tet genes. A total tet gene match was found between the spleen and intestine samples from heron, mallard, kestrel, and gull. A different tet gene distribution was found between the spleen and intestine samples from a Eurasian teal, showing a tet(L) positive intestine sample vs. a tet (M) positive spleen. For each tet gene amplified, the identity of the amplicon was confirmed by comparison between the sequence obtained and the corresponding sequences from antibiotic-resistant Gram-positive or Gram-negative bacteria in the GenBank database, showing 99–100% nucleotide similarity. One sequence for each of the three tetracycline resistance genes amplified was deposited in the GenBank database under accession numbers OP793879, OP793880, and OP807021.
4. Discussion
Microbes, antimicrobial agents, and AMR genes may be cycled and re-cycled through soil, groundwater, marine water, wild animals, crops, shellfish, and livestock [16]. Moreover, the expansion of urban populations and the changes in land use resulting in a fragmentation or loss of natural habitats can lead to the overcoming of natural barriers between humans and wildlife, increasing direct and indirect contact of wildlife with humans and their livestock and potentially expanding the role of wildlife in AMR propagation. Therefore, increasing interest has turned to wild animals in order to evaluate their role as an additional source of antimicrobial resistance.
Antimicrobial resistance has been reported in a wide range of wild animals, such as wild boars [17], wild rabbits [18], wolves [19], lynxes [20], red foxes [21], roe deer [4,22], wild rodents [23], European hedgehogs, and crested porcupines [9].
Wild birds have been speculated to be sentinels, reservoirs, and potential vehicles of resistant bacteria and genetic determinants of AMR [4,8,24,25,26]. In particular, migratory bird species traveling great distances in short periods of time and inhabiting a wide variety of environments could act as efficient AMR dispersers [2,27]. A role for migratory wild birds has also been proposed to explain the occurrence of multidrug-resistant bacteria in places that are isolated from human activities [27].
A characterisation of resistant bacteria isolated from wild birds showed a variety of antimicrobial resistance patterns, including tetracycline resistance [5,10,28,29,30,31,32,33,34].
The extensive use of tetracyclines in clinical practice and their incorporation into livestock feed at subtherapeutic doses as growth promoters, until 2006 when this practice was stopped in the EU [35], has subjected bacterial populations to selection pressure and increased the prevalence of tetracycline resistance, which is considered one of the most abundant antimicrobial resistance mechanisms among pathogenic and commensal microorganisms [36]. Tetracycline resistance is generally caused by the acquisition of tetracycline resistance (tet) genes, often associated with either a mobile plasmid or a transposon that act as vectors, transferring genetic information between bacteria of the same or different species [2,37]. To date, at least 59 tet genes and 11 mosaic tet genes have been described [38]. Three main resistance mechanisms are mediated by tet genes: pumping the drug out of the cell before it reaches its site of action (active efflux pumps), protection of the ribosomal binding site, which decreases drug binding, and enzymatic inactivation of the active compound. The first two mechanisms currently predominate in clinical settings [39].
In this study, we focused on three tet genes representative and frequently detected within each of the three mechanisms of resistance.
In total, 114 (45%) of 254 spleens tested showed one or more tet genes. In particular, tet(M) was detected in 103 (40.5%), tet(L) in 28 (11%), and tet(X) in 4 (1.5%) of the 254 spleen samples. With respect to the 73 intestine samples, 16 (22%) were tet(M) positive, and 4 (5%) were tet(L) positive. No intestine samples tested positive for tet(X). Tet(M) is one of 12 different tet genes that code for proteins that protect bacterial ribosomes from the action of tetracyclines. In particular, it has been proposed that the Tet(M) protein alters the conformation of the tetracycline binding site [40]. The high tet(M) frequency detected in this study, compared to that of the other tet genes tested, was consistent with other reports showing that this gene has the broadest taxonomic distribution among all bacteria, probably because of its association with conjugative chromosomal elements [39]. Conjugative transposons appear to have lower host specificity than plasmids, which may explain the detection of tet(M) in 81 different genera, including 40 Gram-positive and 41 Gram-negative bacteria [41,42]. Tet (L) is one of 33 tet genes that code for energy-dependent efflux of tetracyclines, i.e., the active transport of tetracyclines across the cell membrane. This gene is generally found on small transmissible plasmids. Tet(L) is one of the tet genes that in recent years has shown the largest increase in its distribution among bacterial genera. To date, it has been detected in 25 Gram-positive and 23 Gram-negative bacteria [41,42]. Tet(X) is one of the 13 genes encoding for enzymes that chemically modify tetracycline. In particular, the tet(X) gene encodes a NADP-dependent monooxygenase that catalyses the degradation of tetracycline antimicrobials [43], including tigecycline [44], a next-generation tetracycline that is considered as a last resort antimicrobial against severe infections by pan-drug-resistant bacterial pathogens [45]. This gene was not only observed in obligate anaerobes but also in a variety of environmental aerobic bacteria, as well as diverse human pathogens [45]. Until now, tet(X) has only been found in Gram-negative genera (n. 25), except the genus Clostridium [41,42].
The gene frequency observed in this study agreed with previous reports [41,42]. Tetracycline resistance genes are not randomly distributed among bacteria, but they occur with a different frequency depending the bacterial species and/or genera [46], directing the choice of the target genes on the basis of the bacterial isolates obtained. Otherwise, if culture-independent methods are applied, it would be advisable to test a panel of genes including those with the highest frequency, to avoid the risk of underestimation.
The high tet gene prevalence observed in magpies (71%) and hooded crows (62.5%) is not surprising, considering that these omnivorous and opportunistic species, as well as pigeons and herons, developed a marked synanthropic temperament, taking advantage of the presence of crops, waste, landfills, and all that derives from human activities.
The results observed in waterfowl species, including migratory species, and birds of prey agree with previous studies suggesting contamination via water sources where wastewater from farms and cities is discharged [47,48] or via their prey, such as small wild animals and birds with diverse habitat ranges, including urban and rural environments [49,50]. In this regard, Kozak et al. [51] reported that small wild mammals living in the proximity of farms are generally more likely to harbour resistant bacteria than wild mammals living in natural areas. Predatory birds can also feed on carcasses of livestock animals that can carry more antimicrobial resistant bacterial strains [33]. From an epidemiological perspective, the detection of resistance genes in migratory bird species, as well as in prey birds foraging across large distances, is of particular interest regarding their potential ability to disperse resistance genes or antimicrobial-resistant bacteria over large areas [14], accelerating the globalization of AMR [52].
Antimicrobial resistance genes are a normal finding in the commensal gut microbiota [27]. Considering that the molecular approach did not allow us to associate tet genes with bacterial species, the detection of tet genes from the intestine did not allow us to distinguish pathogenic from commensal bacterial source, whereas the amplification of tet genes from the spleen could be more suggestive of their association with bacteria responsible for infection in the animals tested. Contamination could be excluded because only intact intestines were examined, and DNA extraction was performed by removing an internal fragment from each organ with a disposable scalpel blade. Moreover, nine birds showing tet positive spleens had tet negative intestines.
Previous studies reported the presence of antimicrobial-resistant bacteria and resistance genes in wildlife admitted to rehabilitation centres [30,53,54,55]. Baros Jorquera et al. [5], focusing on a wildlife rehabilitation centre built environment as a possible source of antimicrobial-resistant bacteria in hospitalised animals, showed bacterial environmental contamination that did not entirely overlap with the antimicrobial resistance patterns detected in admitted animals. In this study, the tested birds had been collected already dead or dying and then passed quickly through the rehabilitation centre, where they received no antimicrobial treatment. Therefore, it is likely that they carried resistant bacteria before their admission to the centre. Workers in wildlife centres and people who may have professional or other contact with wild birds should consider the risk of transmission of zoonotic antimicrobial-resistant agents or transmissible resistance determinants and take appropriate preventive measures.
5. Conclusions
The acquisition of resistance genes in wild and free-ranging populations is a concern, as this may increase the environmental reservoir of AMR. The results of this study agree with previous reports suggesting the role of wildlife and wild birds in particular as reservoirs, vectors, or bioindicators of resistant bacteria and genetic determinants of antimicrobial resistance in the environment. In the future, this role could become more significant as wildlife, livestock, and people are more frequently in close contact due increased anthropogenic pressure.
Author Contributions
Conceptualisation, A.D.F. and V.V.E.; methodology, A.D.F., D.S. and F.B.; software D.S.; validation, A.D.F.; formal analysis, A.D.F. and V.V.E.; investigation, A.D.F., D.S. F.B., and V.V.E.; resources, A.D.F., F.B. and V.V.E.; data curation, A.D.F., F.B. and V.V.E.; writing—original draft preparation, A.D.F.; writing—review and editing, A.D.F., D.S., F.B. and V.V.E.; supervision, A.D.F. and V.V.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Ethical review and approval were waived for this study because the samples examined were taken from dead birds.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequences generated in this study are available in GenBank under Accession numbers OP793879, OP793880, and OP807021.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Martin, M.F.; Liras, P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 1989, 43, 173–206. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Donato, J.; Huimi Wang, H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Plaza-Rodríguez, C.; Alt, K.; Grobbel, M.; Hammerl, J.A.; Irrgang, A.; Szabo, I.; Stingl, K.; Schuh, E.; Wiehle, L.; Pfefferkorn, B.; et al. Wildlife as sentinels of antimicrobial resistance in Germany? Front. Vet. Sci. 2021, 7, 627821. [Google Scholar] [CrossRef]
- Baros Jorquera, C.; Moreno-Switt, A.I.; Sallaberry-Pincheira, N.; Munita, J.M.; Flores Navarro, C.; Tardone, R.; González-Rocha, G.; Singer, R.S.; Bueno, I. Antimicrobial resistance in wildlife and in the built environment in a wildlife rehabilitation center. One Health 2021, 13, 100298. [Google Scholar] [CrossRef]
- Guardabassi, L.; Agerso, Y. Genes homologous to glycopeptide resistance vanA are widespread in soil microbial communities. FEMS Microbiol. Lett. 2006, 259, 221–225. [Google Scholar] [CrossRef]
- Seyfried, E.E.; Newton, R.J.; Rubert, K.F., IV; Pedersen, J.A.; McMahon, K.D. Occurrence of tetracycline resistance genes in aquaculture facilities with varying use of oxytetracycline. Microb. Ecol. 2010, 59, 799–807. [Google Scholar] [CrossRef]
- Blanco-Peña, K.; Esperón, F.; Torres-Mejía, A.M.; de la Torre, A.; de la Cruz, E.; Jiménez-Soto, M. Antimicrobial resistance genes in pigeons from public parks in Costa Rica. Zoonoses Public Health 2017, 64, e23–e30. [Google Scholar] [CrossRef]
- Di Francesco, A.; Renzi, M.; Borel, N.; Marti, H.; Salvatore, D. Detection of tetracycline resistance genes in European hedgehogs (Erinaceus europaeus) and crested porcupines (Hystrix cristata). J. Wildl. Dis. 2020, 56, 219–223. [Google Scholar] [CrossRef]
- Luo, Y.; Tan, L.; Zhang, H.; Bi, W.; Zhao, L.; Wang, X.; Lu, X.; Xu, X.; Sun, R.; Alvarez, P.J.J. Characteristics of wild bird resistomes and dissemination of antibiotic resistance genes in interconnected bird-habitat systems revealed by similarity of blaTEM polymorphic sequences. Environ. Sci. Technol. 2022, 56, 15084–15095. [Google Scholar] [CrossRef]
- Galhano, B.S.P.; Ferrari, R.G.; Panzenhagen, P.; de Jesus, A.C.S.; Conte-Junior, C.A. Antimicrobial resistance gene detection methods for bacteria in animal-based foods: A brief review of highlights and advantages. Microorganisms 2021, 9, 923. [Google Scholar] [CrossRef] [PubMed]
- Sundsfjord, A.; Simonsen, G.S.; Haldorsen, B.C.; Haaheim, H.; Hjelmevoll, S.-O.; Littauer, P.; Dahl, K.H. Genetic methods for detection of antimicrobial resistance. APMIS 2004, 112, 815–837. [Google Scholar] [CrossRef] [PubMed]
- Singer, R.S.; Ward, M.P.; Maldonado, G. Can landscape ecology untangle the complexity of antibiotic resistance? Nat. Rev. Microbiol. 2007, 4, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Vittecoq, M.; Godreuil, S.; Prugnolle, F.; Durand, P.; Brazier, L.; Renaud, N.; Arnal, A.; Aberkane, S.; Jean-Pierre, H.; Gauthier-Clerc, M.; et al. Antimicrobial resistance in wildlife. J. Appl. Ecol. 2016, 53, 519–529. [Google Scholar] [CrossRef]
- Ng, L.K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 2001, 15, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Wellington, E.M.H.; Boxall, A.B.A.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Poeta, P.; Costa, D.; Igrejas, G.; Rojo-Bezares, B.; Sáenz, Y.; Zarazaga, M.; Ruiz-Larrea, F.; Rodrigues, J.; Torres, C. Characterization of vanA-containing Enterococcus faecium isolates carrying Tn5397-like and Tn916/Tn1545-like transposons in wild boars (Sus scrofa). Microb. Drug Resist. 2007, 13, 151–156. [Google Scholar] [CrossRef]
- Figueiredo, N.; Radhouani, H.; Gonçalves, A.; Rodrigues, J.; Carvalho, C.; Igrejas, G.; Poeta, P. Genetic characterization of vancomycin-resistant enterococci isolates from wild rabbits. J. Basic Microbiol. 2009, 49, 491–494. [Google Scholar] [CrossRef]
- Gonçalves, A.; Igrejas, G.; Radhouani, H.; Correia, S.; Pacheco, R.; Santos, T.; Monteiro, R.; Guerra, A.; Petrucci-Fonseca, F.; Brito, F.; et al. Antimicrobial resistance in faecal enterococci and Escherichia coli isolates recovered from Iberian wolf. Let. Appl. Microbiol. 2013, 56, 268–274. [Google Scholar] [CrossRef]
- Gonçalves, A.; Igrejas, G.; Radhouani, H.; Santos, T.; Monteiro, R.; Pacheco, R.; Alcaide, E.; Zorrilla, I.; Serra, R.; Torres, C.; et al. Detection of antibiotic resistant enterococci and Escherichia coli in free range Iberian Lynx (Lynx pardinus). Sci. Total Environ. 2013, 456–457, 115–119. [Google Scholar] [CrossRef]
- Radhouani, H.; Igrejas, G.; Gonçalves, A.; Pacheco, R.; Monteiro, R.; Sargo, R.; Brito, F.; Torres, C.; Poeta, P. Antimicrobial resistance and virulence genes in Escherichia coli and enterococci from red foxes (Vulpes vulpes). Anaerobe 2013, 23, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D.; Wang, J.; Fanning, S.; McMahon, B.J. Antimicrobial resistance in wildlife: Implications for public health. Zoonoses Public Health 2015, 62, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Furness, L.E.; Campbell, A.; Zhang, L.; Gaze, W.H.; McDonald, R.A. Wild small mammals as sentinels for the environmental transmission of antimicrobial resistance. Environ. Res. 2017, 154, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Sato, G.; Oka, C.; Asagi, M.; Ishiguro, N. Detection of conjugative R plasmids conferring chloramphenicol resistance in Escherichia coli isolated from domestic and feral pigeons and crows. Zentralbl. Bakteriol. Orig. A 1978, 241, 407–417. [Google Scholar] [PubMed]
- Bonnedahl, J.; Järhult, J.D. Antibiotic resistance in wild birds. Ups. J. Med. Sci. 2014, 119, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Ahlstron, C.A.; Ramey, A.M.; Woksepp, H.; Bonnedahl, J. Repeated detection of carbapenemase-producing Escherichia coli in gulls inhabiting Alaska. Antimicrob. Agents Chemother. 2019, 63, e00758-19. [Google Scholar]
- Arnold, K.E.; Williams, N.J.; Bennett, M. ‘Disperse abroad in the land’: The role of wildlife in the dissemination of antimicrobial resistance. Biol. Lett. 2016, 12, 20160137. [Google Scholar] [CrossRef]
- Livermore, D.M.; Warner, M.; Hall, L.M.; Enne, V.I.; Projan, S.J.; Dunman, P.M.; Wooster, S.L.; Harrison, G. Antibiotic resistance in bacteria from magpies (Pica pica) and rabbits (Oryctolagus cuniculus) from west Wales. Environ. Microbiol. 2001, 3, 658–661. [Google Scholar] [CrossRef]
- Shobrak, M.Y.; Abo-Amer, A.E. Role of wild birds as carriers of multi-drug resistant Escherichia coli and Escherichia vulneris. Braz. J. Microbiol. 2015, 4, 1199–1209. [Google Scholar] [CrossRef]
- Giacopello, C.; Foti, M.; Mascetti, A.; Grosso, F.; Ricciardi, D.; Fisichella, V.; Lo Piccolo, F. Antimicrobial resistance patterns of Enterobacteriaceae in European wild bird species admitted in a wildlife rescue centre. Vet. It. 2016, 52, 139–144. [Google Scholar]
- Guyomard-Rabenirina, S.; Reynaud, Y.; Pot, M.; Albina, E.; Couvin, D.; Ducat, C.; Gruel, G.; Ferdinand, S.; Legreneur, P.; Le Hello, S.; et al. Antimicrobial resistance in wildlife in Guadeloupe (French West Indies): Distribution of a single blaCTX–M–1/IncI1/ST3 plasmid among humans and wild animals. Front. Microbiol. 2020, 11, 1524. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.W.; Choe, J.C.; Jablonski, P.G.; Lee, S. Detection of antibiotic-resistant Escherichia coli from the feces of the Orientalmagpie nestlings. Kor. J. Orni. 2020, 27, 3–9. [Google Scholar] [CrossRef]
- Gambino, D.; Vicari, D.; Vitale, M.; Schirò, G.; Mira, F.; Giglia, M.; Riccardi, A.; Gentile, A.; Giardina, S.; Carrozzo, A.; et al. Study on Bacteria Isolates and Antimicrobial Resistance in Wildlife in Sicily, Southern Italy. Microorganisms 2021, 9, 203. [Google Scholar] [CrossRef] [PubMed]
- Martín-Maldonado, B.; Rodríguez-Alcázar, P.; Fernández-Novo, A.; González, F.; Pastor, N.; López, I.; Suárez, L.; Moraleda, V.; Aranaz, A. Urban birds as antimicrobial resistance sentinels: White storks showed higher multidrug-resistant Eschrichia coli levels than seagulls in central Spain. Animals 2022, 12, 2714. [Google Scholar] [CrossRef]
- Regulation (EC) No 1831/2003 of the European Parliament and of the Council of 22 September 2003 on Additives for Use in Animal Nutrition (Text with EEA Relevance). Available online: http://data.europa.eu/eli/reg/2003/1831/oj (accessed on 3 November 2022).
- Wang, S.; Gao, X.; Gao, Y.; Li, Y.; Cao, M.; Xi, Z.; Zhao, L.; Feng, Z. Tetracycline resistance genes identified from distinct soil environments in China by functional metagenomics. Front. Microbiol. 2017, 8, 1406. [Google Scholar] [CrossRef]
- Hiltunen, T.; Virta, M.; Laine, A.L. Antibiotic resistance in the wild: An eco-evolutionary perspective. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 19, 372. [Google Scholar] [CrossRef]
- Roberts, M.C. Mechanism of Resistance for Characterized tet and otr Genes. 2020. Available online: http://faculty.washington.edu/marilynr/tetweb1.pdf (accessed on 29 October 2022).
- Roberts, M.C. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 2005, 245, 195–203. [Google Scholar] [CrossRef]
- Dönhöfer, A.; Franckenberg, S.; Wickles, S.; Berninghausen, O.; Beckmann, R.; Wilson, D.N. Structural basis for TetM-mediated tetracycline resistance. Proc. Natl. Acad. Sci. USA. 2012, 109, 16900–16905. [Google Scholar] [CrossRef]
- Roberts, M.C. Distribution of tet Resistance Genes among Gram-Positive Bacteria, Mycobacterium, Mycoplasma, Nocardia, Streptomyces and Ureaplasma. Available online: https://faculty.washington.edu/marilynr/tetweb3.pdf (accessed on 29 October 2022).
- Roberts, M.C. Distribution of tet Resistance Genes among Gram-Negative Bacteria. Available online: https://faculty.washington.edu/marilynr/tetweb2.pdf (accessed on 29 October 2022).
- Yang, W.; Moore, I.F.; Koteva, K.P.; Bareich, D.C.; Hughes, D.W.; Wright, G.D. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J. Biol. Chem. 2004, 279, 52346–52352. [Google Scholar] [CrossRef]
- Moore, I.F.; Hughes, D.W.; Wright, G.D. Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry 2005, 44, 11829–11835. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.-X.; Chen, C.; Cui, C.-Y.; Li, X.-P.; Zhang, Y.; Liao, X.-P.; Sun, J.; Liu, Y.-H. Emerging high-level tigecycline resistance: Novel tetracycline destructases spread via the mobile Tet(X). BioEssays 2020, 42, 2000014. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C.; Schwarz, S. Tetracycline and phenicol resistance genes and mechanisms: Importance for agriculture, the environment, and humans. J. Environ. Qual. 2016, 45, 576–592. [Google Scholar] [CrossRef] [PubMed]
- Cole, D.; Drum, D.J.; Stalknecht, D.E.; White, D.G.; Lee, M.D.; Ayers, S.; Sobsey, M.; Maurer, J.J. Free-living Canada geese and antimicrobial resistance. Emerg. Infect. Dis. 2005, 11, 935–938. [Google Scholar] [CrossRef] [PubMed]
- Dolejska, M.; Cizek, A.; Literak, I. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from black-headed gulls in the Czech Republic. J. Appl. Microbiol. 2007, 103, 11–19. [Google Scholar] [CrossRef]
- Marrow, J.; Whittington, J.K.; Mitchell, M.; Hoyer, L.L.; Maddox, C. Prevalence and antibiotic-resistance characteristics of Enterococcus spp. Isolated from free-living and captive raptors in central Illinois. J. Wildl. Dis. 2009, 45, 302–313. [Google Scholar] [CrossRef]
- Guenther, S.; Grobbel, M.; Lübke-Becker, A.; Goedecke, A.; Friedrich, N.D.; Wieler, L.H.; Ewers, C. Antimicrobial resistance profiles of Escherichia coli from common European wild bird species. Vet. Microbiol. 2010, 29, 219–225. [Google Scholar] [CrossRef]
- Kozak, G.K.; Boerlin, P.; Janecko, N.; Reid-Smith, R.J.; Jardine, C. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 2009, 75, 559–566. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef]
- Fuentes-Castillo, D.; Farfán-López, M.; Esposito, F.; Moura, Q.; Fernandes, M.R.; Lopes, R.; Cardoso, B.; Muñoz, M.E.; Cerdeira, L.; Najle, I.; et al. Wild owls colonized by international clones of extended-spectrum β-lactamase (CTX-M)-producing Escherichia coli and Salmonella infantis in the Southern Cone of America. Sci. Total Environ. 2019, 674, 554–562. [Google Scholar] [CrossRef]
- Darwich, L.; Vidal, A.; Seminati, C.; Albamonte, A.; Casado, A.; López, F.; Molina-López, R.A.; Migura-Garcia, L. High prevalence and diversity of extended-spectrum β-lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLoS ONE 2019, 14, e0210686. [Google Scholar] [CrossRef]
- Casalino, G.; D’Amico, F.; Dinardo, F.R.; Bozzo, G.; Napoletano, V.; Camarda, A.; Bove, A.; Lombardi, R.; D’Onghia, F.P.; Circella, E. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli in wild birds from a wildlife rescue centre. Animals 2022, 12, 2889. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).