Associations between Milk Fatty Acid Profile and Body Condition Score, Ultrasound Hepatic Measurements and Blood Metabolites in Holstein Cows
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Data, Milk and Blood Sampling
2.2. Indicators of Metabolic Stress
2.2.1. Ultrasonographic Measurement Acquisition
2.2.2. Hematochemical Parameters Analysis
2.2.3. Body Condition Score
2.3. Milk Fatty Acid Analysis
2.3.1. Sample Preparation and Lipid Extraction
2.3.2. Gas Chromatography
2.4. Statistical Analysis
2.4.1. Multivariate Factor Analysis
2.4.2. Mixed Model Analysis
3. Results
3.1. Metabolic Indicators and Latent Factors of Milk Fatty Acids
3.2. Individual Sources of Variation
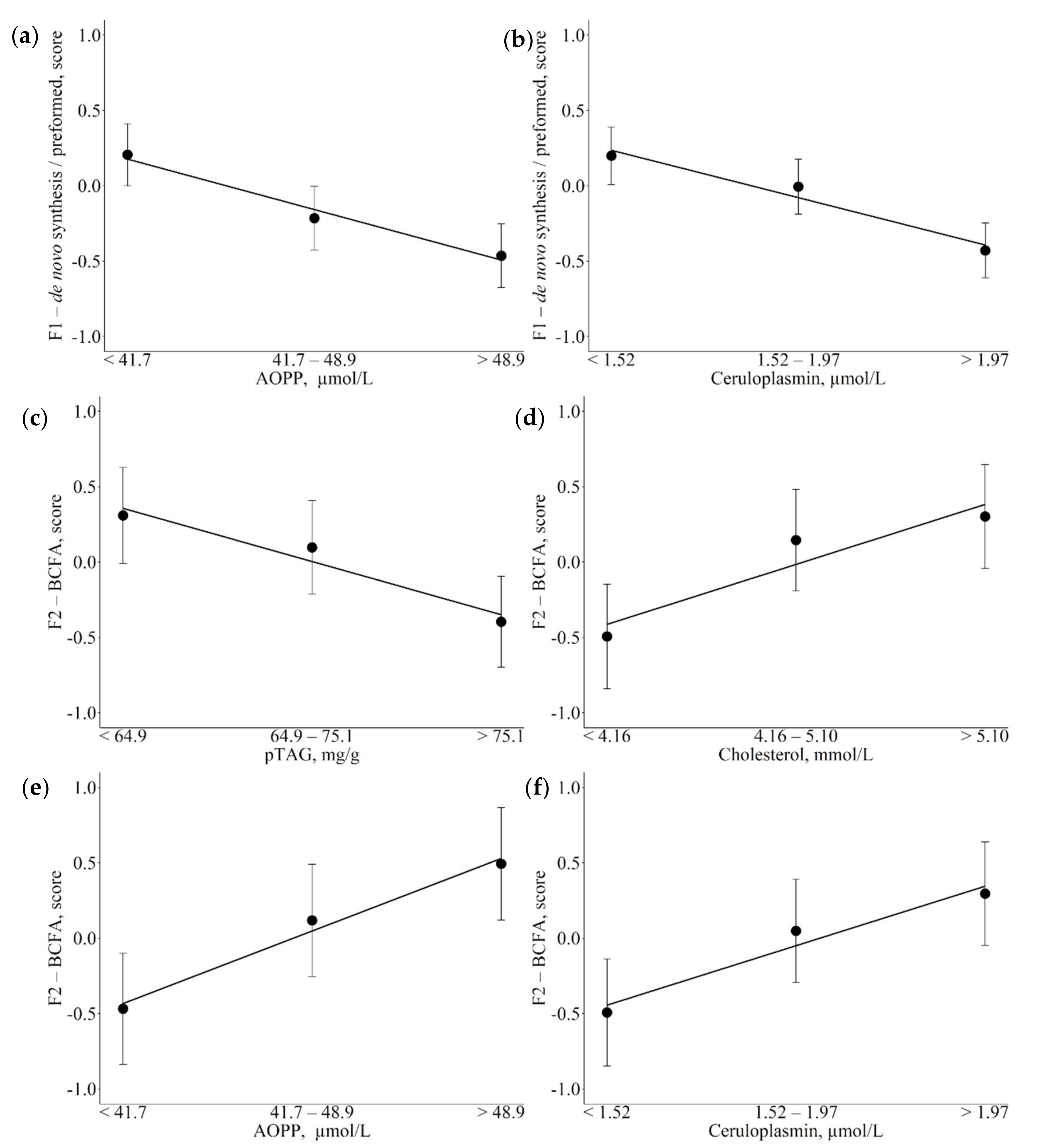
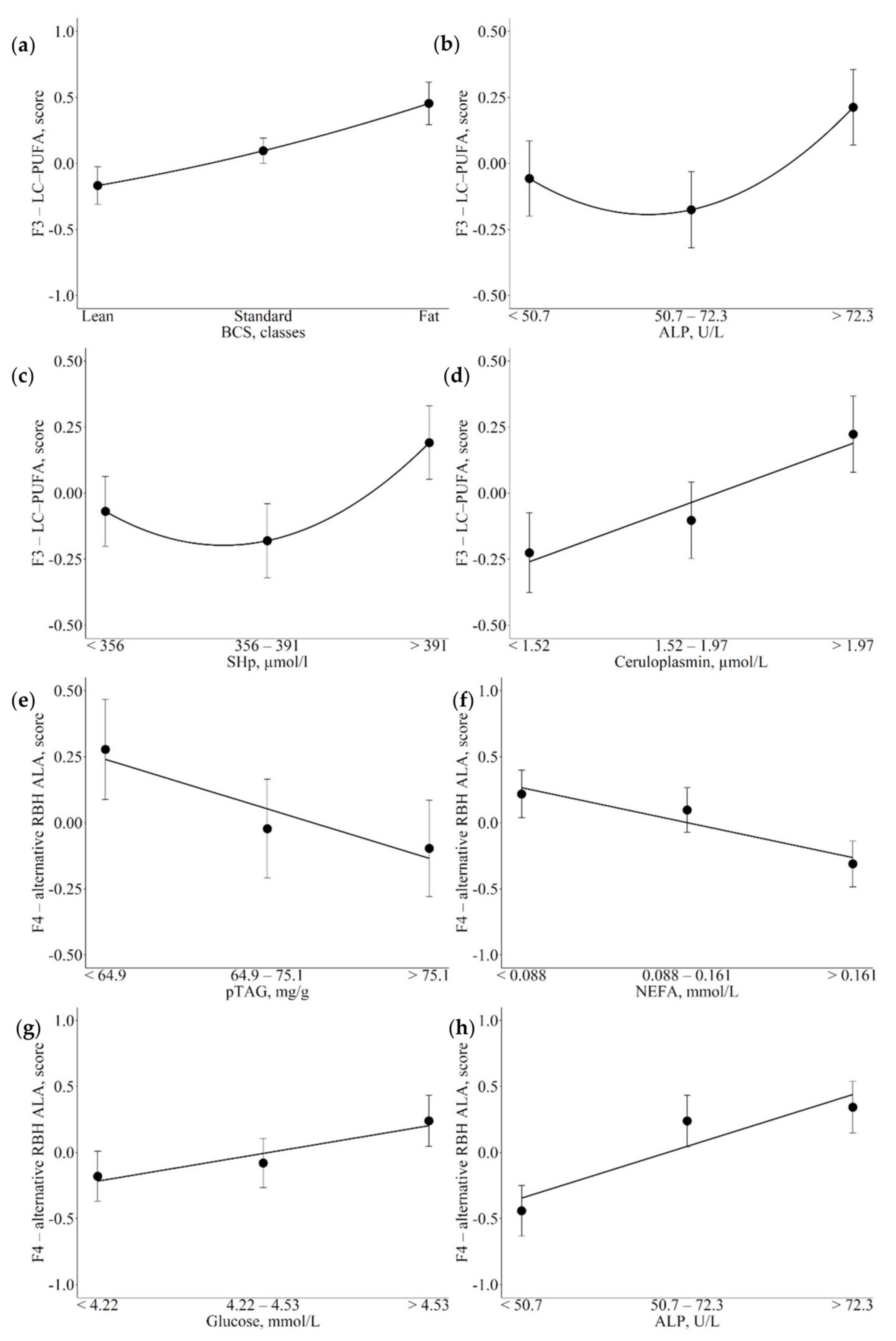
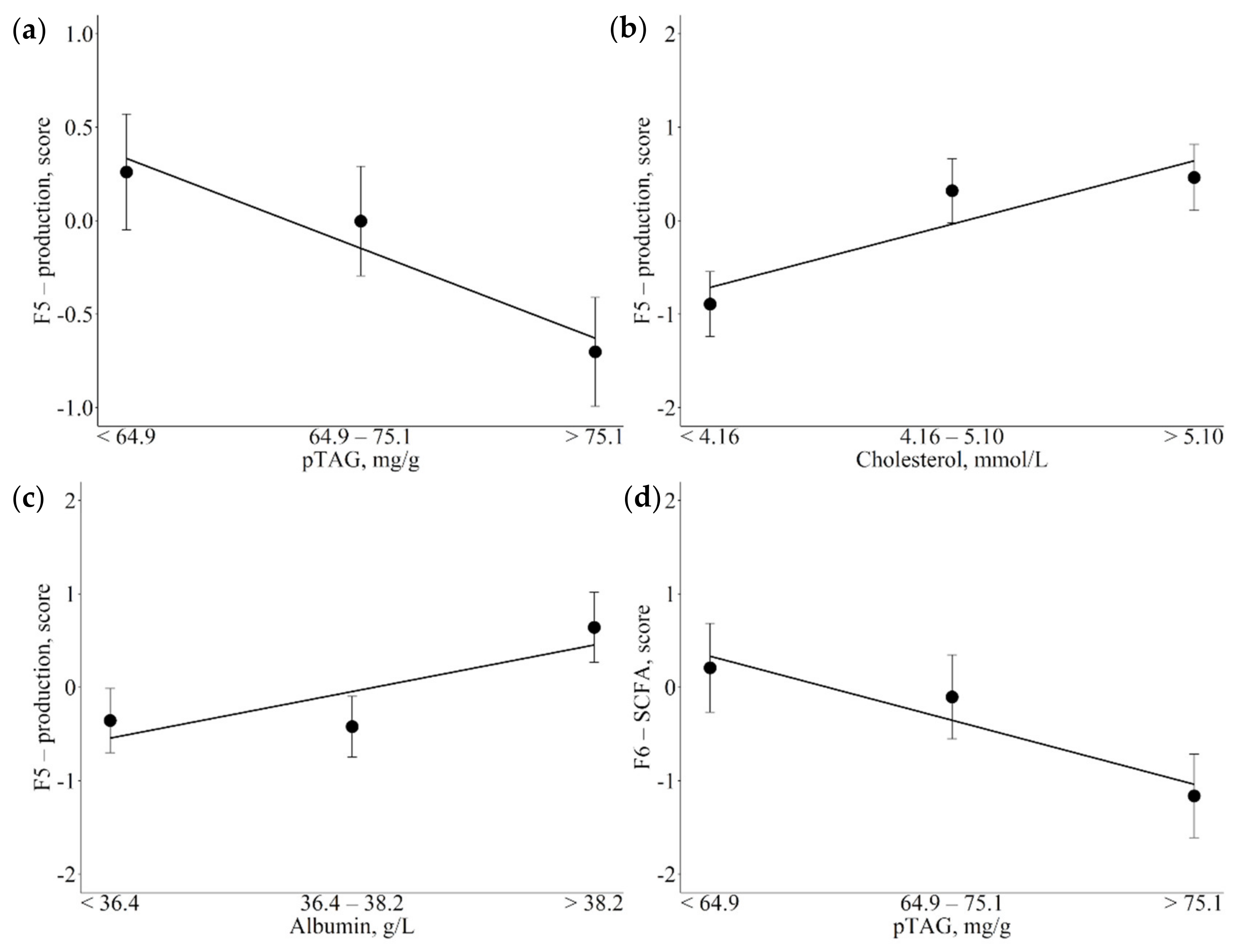
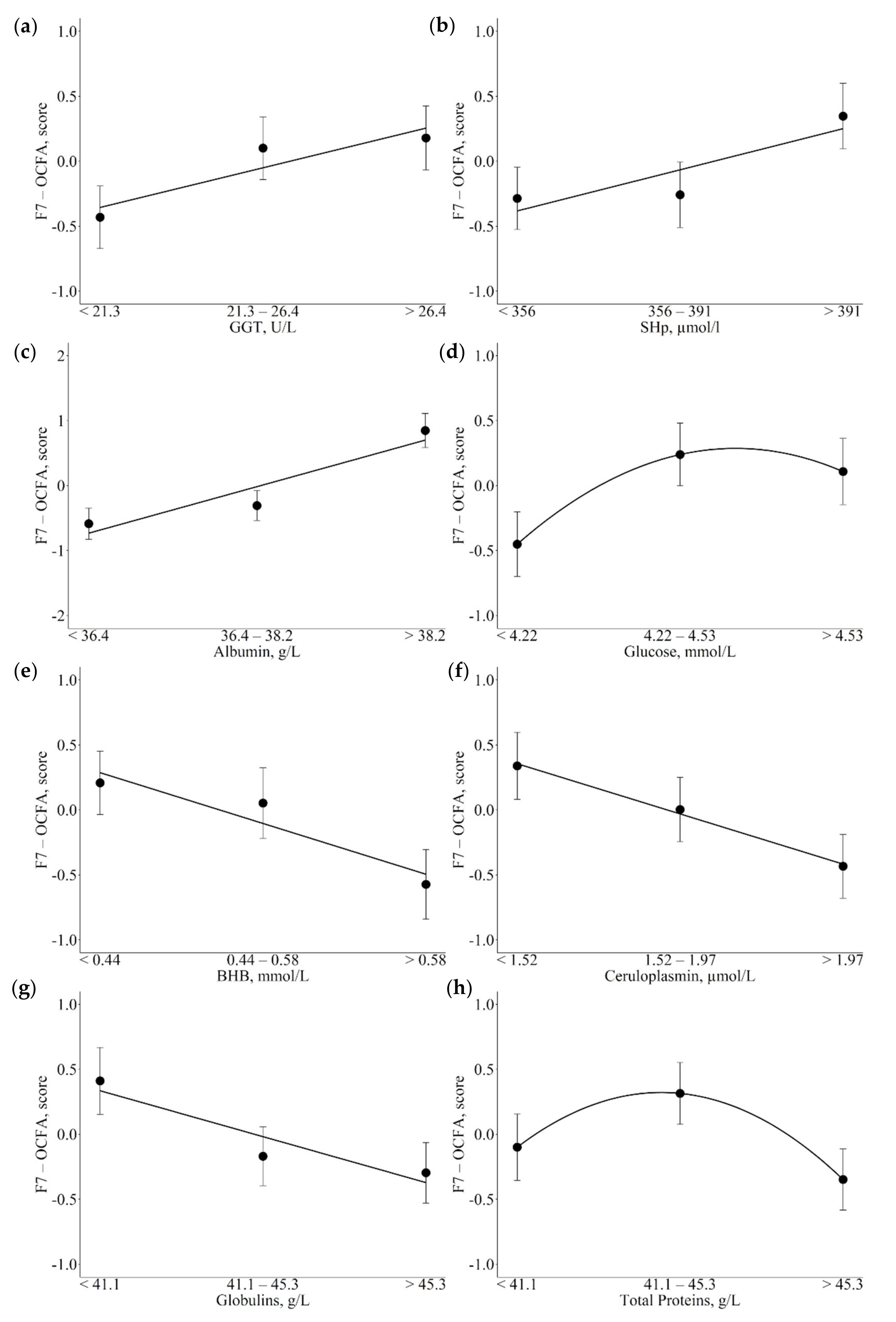
3.3. Associations between Metabolic Indicators and Factors
4. Discussion
4.1. Latent Factors Identify Clusters of Fatty Acids Acting in Common Metabolic Pathways
4.2. Associations of Metabolic Indicators with Fatty Acid Latent Factors Reveal Their Role as Potential Markers of Metabolic Variations
4.3. Indicators of Energy Metabolism
4.3.1. Cholesterol
4.3.2. Glucose, NEFA, and BHB
4.3.3. pTAG
4.3.4. BCS
4.4. Blood Metabolites Related to Inflammation
4.4.1. Ceruloplasmin
4.4.2. Globulin and Protein Serum Levels
4.5. Hepatic Enzymes and Antioxidants
4.5.1. GGT
4.5.2. ALP
4.5.3. AOPP and SHp
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sundrum, A. Metabolic Disorders in the Transition Period Indicate that the Dairy Cows’ Ability to Adapt is Overstressed. Animals 2015, 5, 978–1020. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, F.; Doherty, M. Production diseases of the transition cow. Vet. J. 2008, 176, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Trevisi, E.; Minuti, A. Assessment of the innate immune response in the periparturient cow. Res. Vet. Sci. 2018, 116, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Mezzetti, M.; Bionaz, M.; Trevisi, E. Interaction between inflammation and metabolism in periparturient dairy cows. J. Anim. Sci. 2020, 98, S155–S174. [Google Scholar] [CrossRef] [PubMed]
- Giannuzzi, D.; Tessari, R.; Pegolo, S.; Fiore, E.; Gianesella, M.; Trevisi, E.; Ajmone Marsan, P.; Premi, M.; Piccioli-Cappelli, F.; Tagliapietra, F.; et al. Associations between ultrasound measurements and hematochemical parameters for the assessment of liver metabolic status in Holstein-Friesian cows. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Roche, J.; Kay, J.K.; Friggens, N.; Loor, J.; Berry, D. Assessing and managing body condition score for the prevention of metabolic disease in dairy cows. Vet. Clin. N. Am. Food Anim. Pract. 2013, 29, 323–326. [Google Scholar] [CrossRef]
- Haudum, A.; Starke, A.; Beyerbach, M.; Wohlsein, P.; Rehage, J. Ultrasonographic Assessment of Liver Dimensions in Dairy Cows with Different Hepatic Triacylglycerol Content. J. Anim. Sci. 2011, 89, 1392–1400. [Google Scholar] [CrossRef]
- Starke, A.; Schmidt, S.; Haudum, A.; Scholbach, T.; Wohlsein, P.; Beyerbach, M.; Rehage, J. Evaluation of Portal Blood Flow Using Transcutaneous and Intraoperative Doppler Ultrasonography in Dairy Cows with Fatty Liver. J. Dairy Sci. 2011, 94, 2964–2971. [Google Scholar] [CrossRef] [Green Version]
- Banzato, T.; Fiore, E.; Morgante, M.; Manuali, E.; Zotti, A. Texture Analysis of B-mode Ultrasound Images to Stage Hepatic Lipidosis in the Dairy Cow: A Methodological Study. Res. Vet. Sci. 2016, 108, 71–75. [Google Scholar] [CrossRef]
- Peng, Z.; Li, X.; Wang, Z.; Liu, G.; Li, X. The effects of non-esterified fatty acids and β-hydroxybutyrate on the hepatic CYP2E1 in cows with clinical ketosis. J. Dairy Res. 2019, 86, 68–72. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, B.; Shu, S.; Zhang, H.; Xu, C.; Wu, L.; Xia, C. Critical thresholds of liver function parameters for ketosis prediction in dairy cows using receiver operating characteristic (ROC) analysis. Vet. Q. 2015, 35, 159–164. [Google Scholar] [CrossRef]
- Du, X.; Chen, L.; Huang, D.; Peng, Z.; Zhao, C.; Zhang, Y.; Zhu, Y.; Wang, Z.; Li, X.; Liu, G. Elevated Apoptosis in the Liver of Dairy Cows with Ketosis. Cell. Physiol. Biochem. 2017, 43, 568–578. [Google Scholar] [CrossRef]
- Overton, T.; McArt, J.; Nydam, D. A 100-Year Review: Metabolic health indicators and management of dairy cattle. J. Dairy Sci. 2017, 100, 10398–10417. [Google Scholar] [CrossRef] [Green Version]
- Bertoni, G.; Trevisi, E. Use of the Liver Activity Index and Other Metabolic Variables in the Assessment of Metabolic Health in Dairy Herds. Vet. Clin. N. Am. Food Anim. Pract. 2013, 29, 413–431. [Google Scholar] [CrossRef]
- Chilliard, Y.; Ferlay, A.; Mansbridge, R.M.; Doreau, M. Ruminant milk fat plasticity: Nutritional control of saturated, polyunsaturated, trans and conjugated fatty acids. Ann. Zootech. 2000, 49, 181–205. [Google Scholar] [CrossRef] [Green Version]
- Bauman, D.; Griinari, J. Nutritional regulation of milk fat synthesis. Annu. Rev. Nutr. 2003, 23, 203–227. [Google Scholar] [CrossRef] [Green Version]
- Loften, J.; Linn, J.; Drackley, J.; Jenkins, T.; Soderholm, C.; Kertz, A. Invited review: Palmitic and stearic acid metabolism in lactating dairy cows. J. Dairy Sci. 2014, 97, 4661–4674. [Google Scholar] [CrossRef]
- Van Haelst, Y.; Beeckman, A.; Van Knegsel, A.; Fievez, V. Short communication: Elevated concentrations of oleic acid and long-chain fatty acids in milk fat of multiparous subclinical ketotic cows. J. Dairy Sci. 2008, 91, 4683–4688. [Google Scholar] [CrossRef]
- Lock, A.; Bauman, D. Modifying milk fat composition of dairy cows to enhance fatty acids beneficial to human health. Lipids 2004, 39, 1197–1206. [Google Scholar] [CrossRef] [Green Version]
- Mele, M.; Macciotta, N.; Cecchinato, A.; Conte, G.; Schiavon, S.; Bittante, G. Multivariate factor analysis of detailed milk fatty acid profile: Effects of dairy system, feeding, herd, parity, and stage of lactation. J. Dairy Sci. 2016, 99, 9820–9833. [Google Scholar] [CrossRef] [Green Version]
- Palombo, V.; Conte, G.; Mele, M.; Macciotta, N.; Stefanon, B.; Marsan, P.; D’Andrea, M. Use of multivariate factor analysis of detailed milk fatty acid profile to perform a genome-wide association study in Italian Simmental and Italian Holstein. J. Appl. Genet. 2020, 61, 451–463. [Google Scholar] [CrossRef]
- Turini, L.; Conte, G.; Bonelli, F.; Serra, A.; Sgorbini, M.; Mele, M. Multivariate factor analysis of milk fatty acid composition in relation to the somatic cell count of single udder quarters. J. Dairy Sci. 2020, 103, 7392–7406. [Google Scholar] [CrossRef]
- Macciotta, N.; Vicario, D.; Di Mauro, C.; Cappio-Borlino, A. A multivariate approach to modeling shapes of individual lactation curves in cattle. J. Dairy Sci. 2004, 87, 1092–1098. [Google Scholar] [CrossRef]
- Cecchinato, A.; Macciotta, N.; Mele, M.; Tagliapietra, F.; Schiavon, S.; Bittante, G.; Pegolo, S. Genetic and genomic analyses of latent variables related to the milk fatty acid profile, milk composition, and udder health in dairy cattle. J. Dairy Sci. 2019, 102, 5254–5265. [Google Scholar] [CrossRef]
- Fiore, E.; Perillo, L.; Morgante, M.; Giudice, E.; Contiero, B.; Curone, G.; Manuali, E.; Pavone, S.; Piccione, G.; Gianesella, M. Ultrasonographic Measurement of Liver, Portal Vein, Hepatic Vein and Perivisceral Adipose Tissue in High-Yielding Dairy Cows With Fatty Liver During the Transition Period. J. Dairy Res. 2018, 85, 431–438. [Google Scholar] [CrossRef]
- Mezzetti, M.; Minuti, A.; Piccioli-Cappelli, F.; Amadori, M.; Bionaz, M.; Trevisi, E. The role of altered immune function during the dry period in promoting the development of subclinical ketosis in early lactation. J. Dairy Sci. 2019, 102, 9241–9258. [Google Scholar] [CrossRef]
- Edmonson, A.; Lean, I.; Weaver, L.; Farver, T.; Webster, G. A Body Condition Scoring Chart for Holstein Dairy Cows. J. Dairy Sci. 1989, 72, 68–78. [Google Scholar] [CrossRef]
- Piazza, M.; Giannuzzi, D.; Tessari, R.; Fiore, E.; Gianesella, M.; Pegolo, S.; Schiavon, S.; Trevisi, E.; Piccioli Cappelli, F.; Cecchinato, A.; et al. Associations between ultrasound hepatic measurements, body measures and milk production traits in Holstein cows. J. Dairy Sci. 2022, in press. [Google Scholar] [CrossRef]
- Christie, W.W. Preparation of ester derivatives of fatty acids for chromatographic analysis. In Advances in Lipid Methodology Two, 1st ed.; Oily Press: Dundee, UK, 1993; pp. 69–111. [Google Scholar]
- Delmonte, P.; Fardin-Kia, A.; Kramer, J.; Mossoba, M.; Sidisky, L.; Tyburczy, C.; Rader, J. Evaluation of highly polar ionic liquid gas chromatographic column for the determination of the fatty acids in milk fat. J. Chromatogr. A 2012, 1233, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D. Multivariate Statistical Methods; McGraw-Hill: New York, NY, USA, 1976. [Google Scholar]
- Kaiser, H.; Rice, J. Little Jiffy, Mark Iv. Educ. Psychol. Meas. 1974, 34, 111–117. [Google Scholar] [CrossRef]
- Hair, J.F.; Black, W.C.; Babin, B.J.; Anderson, R.E. Multivariate Data Analysis: A Global Perspective, 7th ed.; Pearson Education: London, UK, 2010. [Google Scholar]
- Garnsworthy, P.; Masson, L.; Lock, A.; Mottram, T. Variation of milk citrate with stage of lactation and de novo fatty acid synthesis in dairy cows. J. Dairy Sci. 2006, 89, 1604–1612. [Google Scholar] [CrossRef] [Green Version]
- Vlaeminck, B.; Fievez, V.; Demeyer, D.; Dewhurst, R. Effect of forage:concentrate ratio on fatty acid composition of rumen bacteria isolated from ruminal and duodenal digesta. J. Dairy Sci. 2006, 89, 2668–2678. [Google Scholar] [CrossRef]
- Bas, P.; Archimède, H.; Rouzeau, A.; Sauvant, D. Fatty acid composition of mixed-rumen bacteria: Effect of concentration and type of forage. J. Dairy Sci. 2003, 86, 2940–2948. [Google Scholar] [CrossRef] [Green Version]
- Bionaz, M.; Loor, J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [Green Version]
- Jensen, R. The composition of bovine milk lipids: January 1995 to December 2000. J. Dairy Sci. 2002, 85, 295–350. [Google Scholar] [CrossRef]
- Shingfield, K.; Bernard, L.; Leroux, C.; Chilliard, Y. Role of trans fatty acids in the nutritional regulation of mammary lipogenesis in ruminants. Animal 2010, 4, 1140–1166. [Google Scholar] [CrossRef] [Green Version]
- Vlaeminck, B.; Fievez, V.; Tamminga, S.; Dewhurst, R.; van Vuuren, A.; De Brabander, D.; Demeyer, D. Milk odd- and branched-chain fatty acids in relation to the rumen fermentation pattern. J. Dairy Sci. 2006, 89, 3954–3964. [Google Scholar] [CrossRef] [Green Version]
- Shingfield, K.; Griinari, J. Role of biohydrogenation intermediates in milk fat depression. Eur. J. Lipid Sci. Technol. 2007, 109, 799–816. [Google Scholar] [CrossRef]
- Bionaz, M.; Vargas-Bello-Pérez, E.; Busato, S. Advances in fatty acids nutrition in dairy cows: From gut to cells and effects on performance. J. Anim. Sci. Biotechnol. 2020, 11, 1–36. [Google Scholar] [CrossRef]
- Gross, J.; Schwinn, A.; Müller, E.; Münger, A.; Dohme-Meier, F.; Bruckmaier, R. Plasma cholesterol levels and short-term adaptations of metabolism and milk production during feed restriction in early lactating dairy cows on pasture. J. Anim. Physiol. Anim. Nutr. 2021, 105, 1024–1033. [Google Scholar] [CrossRef]
- Bertoni, G.; Trevisi, E.; Han, X.; Bionaz, M. Effects of inflammatory conditions on liver activity in puerperium period and consequences for performance in dairy cows. J. Dairy Sci. 2008, 91, 3300–3310. [Google Scholar] [CrossRef] [Green Version]
- Drackley, J. Biology of Dairy Cows During the Transition Period: The Final Frontier? J. Dairy Sci. 1999, 82, 2259–2273. [Google Scholar] [CrossRef]
- Grummer, R. Etiology of Lipid-Related Metabolic Disorders in Periparturient Dairy Cows. J. Dairy Sci. 1993, 76, 3882–3896. [Google Scholar] [CrossRef]
- Bobe, G.; Young, J.; Beitz, D. Invited Review: Pathology, Etiology, Prevention, and Treatment of Fatty Liver in Dairy Cows. J. Dairy Sci. 2004, 87, 3105–3124. [Google Scholar] [CrossRef] [Green Version]
- Ametaj, B.; Bradford, B.; Bobe, G.; Nafikov, R.; Lu, Y.; Joung, J.; Beitz, D. Strong relationships between mediators of the acutephase response and fatty liver in dairy cows. Can. J. Anim. Sci. 2005, 85, 165–175. [Google Scholar] [CrossRef]
- Bickerstaffe, R.; Annison, E.; Linzell, J.L. The metabolism of glucose, acetate, lipids and amino acids in lactating dairy cows. J. Agric. Sci. 1974, 82, 71–85. [Google Scholar] [CrossRef]
- Churakov, M.; Karlsson, J.; Edvardsson Rasmussen, A.; Holtenius, K. Milk fatty acids as indicators of negative energy balance of dairy cows in early lactation. Animal 2021, 15, 100253. [Google Scholar] [CrossRef]
- Montgomery, M.; Osborne, B.; Brown, S.; Small, L.; Mitchell, T.; Cooney, G.; Turner, N. Contrasting metabolic effects of medium- versus long-chain fatty acids in skeletal muscle. J. Lipid Res. 2013, 54, 3322–3333. [Google Scholar] [CrossRef] [Green Version]
- Loor, J.; Everts, R.; Bionaz, M.; Dann, H.; Morin, D.; Oliveira, R.; Rodriguez-Zas, S.; Drackley, J.; Lewin, H. Nutrition-induced ketosis alters metabolic and signaling gene networks in liver of periparturient dairy cows. Physiol. Genom. 2007, 32, 105–116. [Google Scholar] [CrossRef] [Green Version]
- Trevisi, E.; Jahan, N.; Bertoni, G.; Ferrari, A.; Minuti, A. Pro-Inflammatory Cytokine Profile in Dairy Cows: Consequences for New Lactation. Ital. J. Anim. Sci. 2015, 14, 3862. [Google Scholar] [CrossRef]
- Venn-Watson, S.; Lumpkin, R.; Dennis, E. Efficacy of dietary odd-chain saturated fatty acid pentadecanoic acid parallels broad associated health benefits in humans: Could it be essential? Sci. Rep. 2020, 10, 8161. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; Stepnowski, P.; Kaska, L.; Proczko, M.; Wisniewski, P.; Sledzinski, M.; Sledzinski, T. A comprehensive study of serum odd- and branched-chain fatty acids in patients with excess weight. Obesity 2016, 24, 1669–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burkhead, J.; Lutsenko, L. The Role of Copper as a Modifier of Lipid Metabolism. In Lipid Metabolism; Baez, R.V., Ed.; IntechOpen: London, UK, 2013. [Google Scholar]
- Abu-Salah, K.; al-Othman, A.; Lei, K. Lipid composition and fluidity of the erythrocyte membrane in copper-deficient rats. Br. J. Nutr. 1992, 68, 435–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taormina, V.; Unger, A.; Schiksnis, M.; Torres-Gonzalez, M.; Kraft, J. Branched-Chain Fatty Acids-An Underexplored Class of Dairy-Derived Fatty Acids. Nutrients 2020, 12, 2875. [Google Scholar] [CrossRef]
- Bobbo, T.; Fiore, E.; Gianesella, M.; Morgante, M.; Gallo, L.; Ruegg, P.; Bittante, G.; Cecchinato, A. Variation in blood serum proteins and association with somatic cell count in dairy cattle from multi-breed herds. Animal 2017, 11, 2309–2319. [Google Scholar] [CrossRef]
- Cattaneo, L.; Lopreiato, V.; Piccioli-Cappelli, F.; Trevisi, E.; Minuti, A. Plasma albumin-to-globulin ratio before dry-off as a possible index of inflammatory status and performance in the subsequent lactation in dairy cows. J. Dairy Sci. 2021, 104, 8228–8242. [Google Scholar] [CrossRef]
- Don, B.; Kaysen, G. Serum albumin: Relationship to inflammation and nutrition. Semin. Dial. 2004, 17, 432–437. [Google Scholar] [CrossRef]
- Trevisi, E.; Amadori, M.; Cogrossi, S.; Razzuoli, E.; Bertoni, G. Metabolic stress and inflammatory response in high-yielding, periparturient dairy cows. Res. Vet. Sci. 2012, 93, 695–704. [Google Scholar] [CrossRef]
- Nishi, K.; Yamasaki, K.; Otagiri, M. Serum Albumin, Lipid and Drug Binding. Subcell. Biochem. 2020, 94, 383–397. [Google Scholar] [CrossRef]
- Kauppinen, K. ALAT, AP, ASAT, GGT, OCT activities and urea and total bilirubin concentrations in plasma of normal and ketotic dairy cows. Zentralblatt für Veterinärmedizin Reihe A 1984, 31, 567–576. [Google Scholar] [CrossRef]
- Premi, M.; Mezzetti, M.; Ferronato, G.; Barbato, M.; Piccioli Cappelli, F.; Minuti, A.; Trevisi, E. Changes of Plasma Analytes Reflecting Metabolic Adaptation to the Different Stages of the Lactation Cycle in Healthy Multiparous Holstein Dairy Cows Raised in High-Welfare Conditions. Animals 2021, 11, 1714. [Google Scholar] [CrossRef]
- Vlaeminck, B.; Gervais, R.; Rahman, M.; Gadeyne, F.; Gorniak, M.; Doreau, M.; Fievez, V. Postruminal synthesis modifies the odd- and branched-chain fatty acid profile from the duodenum to milk. J. Dairy Sci. 2015, 98, 4829–4840. [Google Scholar] [CrossRef]
- Gruffat, D.; Durand, D.; Graulet, B.; Bauchart, D. Regulation of VLDL Synthesis and Secretion in the Liver. Reprod. Nutr. Dev. 1996, 36, 375–389. [Google Scholar] [CrossRef]
- Kerner, A.; Avizohar, O.; Sella, R.; Bartha, P.; Zinder, O.; Markiewicz, W.; Levy, Y.; Brook, G.; Aronson, D. Association between elevated liver enzymes and C-reactive protein: Possible hepatic contribution to systemic inflammation in the metabolic syndrome. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 193–197. [Google Scholar] [CrossRef] [Green Version]
- Rezamand, P.; McGuire, M. Effects of trans fatty acids on markers of inflammation in bovine mammary epithelial cells. J. Dairy Sci. 2011, 94, 316–320. [Google Scholar] [CrossRef] [Green Version]
- Gabai, G.; De Luca, E.; Miotto, G.; Zin, G.; Stefani, A.; Da Dalt, L.; Barberio, A.; Celi, P. Relationship between Protein Oxidation Biomarkers and Uterine Health in Dairy Cows during the Postpartum Period. Antioxidants 2019, 8, 21. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Tang, W.; Zhao, M.; Gong, B.; Cao, M.; Li, J. Study on the regulation mechanism of lipopolysaccharide on oxidative stress and lipid metabolism of bovine mammary epithelial cells. Physiol. Res. 2021, 70, 777–785. [Google Scholar] [CrossRef]
- Mollica, M.; Trinchese, G.; Cimmino, F.; Penna, E.; Cavaliere, G.; Tudisco, R.; Musco, N.; Manca, C.; Catapano, A.; Monda, M.; et al. Milk Fatty Acid Profiles in Different Animal Species: Focus on the Potential Effect of Selected PUFAs on Metabolism and Brain Functions. Nutrients 2021, 13, 1111. [Google Scholar] [CrossRef]
- Richard, D.; Kefi, K.; Barbe, U.; Bausero, P.; Visioli, F. Polyunsaturated fatty acids as antioxidants. Pharmacol. Res. 2008, 57, 451–455. [Google Scholar] [CrossRef]
| Herd A 1 | Herd B 1 | ||
|---|---|---|---|
| Lactation | Lactation Primiparous | Lactation Multiparous | |
| Corn silage | 12.42 | 30.76 | 32.94 |
| Sorghum silage | 24.6 | - | - |
| Wheat silage | - | 9.49 | 9.49 |
| Alfalfa hay | 20.98 | 11.47 | 11.48 |
| Ryegrass hay | 2.12 | - | - |
| Barley straw | - | - | - |
| Corn grain ground | 12.62 | 23.95 | 21.16 |
| Barley grain ground | 8.41 | - | - |
| Soybean meal (solvent ex. 44% CP) | 12.1 | 9.25 | 10.65 |
| Sunflower meal (solvent ex. 25% CP) | 4.34 | 4.69 | 4.43 |
| Soybean, whole | - | 3.52 | 3.32 |
| Wheat middlings | - | 4.58 | 4.32 |
| Hydrogenated fat | 0.78 | 0.55 | 0.52 |
| Minerals and vitamin supplements 1 | 1.63 | 1.76 | 1.69 |
| Nutrient composition | |||
| UFL (U/kg DM) | 0.97 | 0.98 | 0.98 |
| ENl (Mcal/kg DM) | 1.55 | 1.59 | 1.53 |
| Crude protein (% DM) | 16.50 | 15.97 | 16.34 |
| Metabolizable protein (% DM) | 10.50 | 10.79 | 10.92 |
| NSC (% DM) | 36.80 | 44.07 | 43.06 |
| NDF (% DM) | 35.80 | 32.18 | 32.91 |
| Item 1 | F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 |
|---|---|---|---|---|---|---|---|---|
| De Novo/ Preformed | BCFA 2 | LC-PUFA 3 | alt RBH ALA 4 | Production | SCFA 5 | OCFA 6 | RBH 7 | |
| Milk yield, kg | 0.12 | −0.21 | −0.07 | −0.12 | 0.92 | −0.04 | 0.03 | 0.06 |
| Milk composition, kg | ||||||||
| Fat | 0.01 | −0.12 | −0.11 | −0.04 | 0.82 | −0.03 | −0.03 | −0.12 |
| Protein | 0.26 | −0.28 | −0.02 | −0.04 | 0.82 | −0.04 | 0.08 | 0.09 |
| Individual fatty acids, g/100 g of total FAs | ||||||||
| SFA | ||||||||
| 4:0 | −0.19 | 0.26 | −0.13 | 0.00 | 0.00 | 0.65 | −0.22 | −0.11 |
| 6:0 | 0.31 | 0.30 | −0.21 | 0.12 | −0.04 | 0.75 | −0.07 | −0.16 |
| 8:0 | 0.62 | 0.27 | −0.12 | 0.20 | −0.01 | 0.65 | 0.03 | −0.12 |
| 10:0 | 0.83 | 0.17 | 0.02 | 0.17 | 0.03 | 0.37 | 0.13 | −0.08 |
| 11:0 | 0.54 | −0.30 | 0.08 | 0.14 | −0.01 | 0.03 | 0.71 | 0.06 |
| 12:0 | 0.89 | 0.12 | 0.04 | 0.12 | 0.03 | 0.18 | 0.19 | −0.06 |
| 13:0 iso | 0.09 | 0.67 | −0.03 | 0.11 | −0.11 | −0.04 | −0.17 | −0.27 |
| 13:0 | 0.68 | −0.22 | 0.14 | 0.07 | 0.04 | −0.13 | 0.60 | 0.06 |
| 14:0 iso | 0.20 | 0.75 | 0.19 | 0.02 | −0.14 | 0.16 | −0.12 | −0.24 |
| 14:0 | 0.81 | 0.12 | −0.25 | 0.03 | 0.11 | 0.06 | 0.09 | −0.09 |
| 15:0 iso | −0.09 | 0.88 | −0.03 | 0.13 | −0.10 | 0.07 | −0.07 | −0.14 |
| 15:0 ante | 0.03 | 0.82 | 0.03 | 0.05 | −0.17 | 0.08 | 0.03 | 0.06 |
| 15:0 | 0.49 | −0.19 | −0.05 | 0.09 | 0.04 | −0.17 | 0.76 | 0.08 |
| 16:0 iso | −0.19 | 0.64 | 0.09 | −0.09 | −0.17 | 0.26 | −0.32 | −0.09 |
| 17:0 iso | −0.66 | 0.45 | −0.02 | 0.00 | −0.04 | 0.11 | −0.07 | 0.18 |
| 17:0 ante | −0.68 | 0.36 | 0.03 | −0.27 | −0.07 | 0.28 | −0.11 | 0.28 |
| 17:0 | −0.71 | −0.07 | −0.22 | −0.14 | −0.08 | 0.27 | 0.37 | 0.16 |
| 18:0 | −0.61 | 0.31 | −0.28 | 0.43 | −0.19 | 0.16 | −0.22 | 0.01 |
| MUFA | ||||||||
| 10:1 (c9) | 0.85 | 0.13 | 0.19 | 0.00 | 0.07 | −0.02 | 0.01 | −0.17 |
| 14:1 (c9) | 0.63 | −0.18 | 0.33 | −0.24 | 0.20 | −0.37 | 0.03 | −0.07 |
| 16:1 (t9) | −0.73 | 0.00 | 0.32 | −0.25 | 0.01 | 0.07 | −0.04 | 0.17 |
| 18:1 (t6+t8) | −0.31 | −0.03 | 0.14 | 0.08 | 0.01 | −0.05 | −0.03 | 0.64 |
| 18:1 (t9) | −0.15 | −0.06 | 0.19 | −0.05 | −0.01 | −0.11 | 0.01 | 0.88 |
| 18:1 (t10) | −0.11 | −0.33 | 0.16 | −0.03 | 0.03 | −0.01 | 0.19 | 0.65 |
| 18:1 (t11) | −0.32 | 0.05 | 0.19 | −0.03 | −0.01 | 0.03 | 0.02 | 0.80 |
| 18:1 (c9) | −0.76 | −0.17 | 0.41 | −0.06 | 0.00 | −0.18 | −0.21 | 0.05 |
| 18:1 (c12) | 0.16 | −0.26 | 0.38 | 0.33 | 0.15 | −0.04 | 0.11 | 0.64 |
| 18:1 (t16 + c14) | 0.06 | 0.03 | 0.07 | 0.68 | 0.04 | −0.14 | 0.07 | 0.42 |
| PUFA | ||||||||
| 18:2 (c9, c12) | −0.08 | −0.22 | 0.84 | −0.01 | −0.04 | −0.1 | −0.09 | 0.21 |
| 18:3 (c9, c12, c15) | 0.04 | −0.07 | 0.75 | 0.07 | −0.02 | 0.01 | 0.05 | 0.21 |
| 18:2 (c9, t11) | −0.07 | 0.07 | 0.76 | −0.12 | 0.03 | −0.22 | −0.06 | 0.2 |
| 20:3 (c8, c11, c14) | 0.19 | 0.06 | 0.63 | 0.20 | 0.07 | −0.16 | 0.03 | −0.01 |
| 20:4 (c5, c8, c11, c14) | −0.09 | −0.10 | 0.74 | 0.11 | −0.12 | −0.01 | 0.09 | 0.1 |
| Eigenvalue | 8.89 | 5.10 | 4.92 | 2.35 | 2.76 | 2.66 | 2.31 | 3.98 |
| Traits 1 | F1-De Novo/Preformed | F2-BCFA 2 | F3-LC-PUFA 3 | F4-alt RBH ALA 4 | ||||
|---|---|---|---|---|---|---|---|---|
| p-value | RMSE 5 | p-Value | RMSE 5 | p-Value | RMSE 5 | p-Value | RMSE 5 | |
| Individual sources of variation 6 | ||||||||
| DIM, classes 7 | <0.001 | 0.931 | 0.069 | 1.244 | 0.098 | 0.655 | 0.126 | 0.675 |
| Parity, classes 8 | 0.990 | 0.362 | 0.979 | 0.249 | ||||
| Body measure | ||||||||
| BCS, classes | 0.764 | 0.932 | 0.278 | 1.261 | 0.007 | 0.649 | 0.229 | 0.650 |
| Ultrasound measurements | ||||||||
| pTAG, mg/g | 0.199 | 0.926 | 0.028 | 1.252 | 0.120 | 0.661 | 0.025 | 0.667 |
| PVA, mm2 | 0.106 | 0.906 | 0.336 | 1.261 | 0.128 | 0.663 | 0.997 | 0.681 |
| PVD, mm | 0.816 | 0.928 | 0.874 | 1.283 | 0.418 | 0.668 | 0.828 | 0.684 |
| LD, mm | 0.809 | 0.941 | 0.668 | 1.302 | 0.314 | 0.663 | 0.698 | 0.680 |
| Hematochemical parameters | ||||||||
| Hematocrit, L/L | 0.972 | 0.927 | 0.811 | 1.308 | 0.050 | 0.679 | 0.346 | 0.666 |
| Energy-related metabolites | ||||||||
| Glucose, mmol/L | 0.926 | 0.924 | 0.538 | 1.303 | 0.204 | 0.687 | 0.019 | 0.649 |
| Cholesterol, mmol/L | 0.398 | 0.922 | 0.046 | 1.292 | 0.504 | 0.687 | 0.667 | 0.667 |
| NEFA, mmol/L | 0.109 | 0.914 | 0.928 | 1.311 | 0.473 | 0.688 | 0.015 | 0.660 |
| BHB, mmol/L | 0.597 | 0.927 | 0.46 | 1.308 | 0.373 | 0.689 | 0.233 | 0.668 |
| Urea, mmol/L | 0.179 | 0.917 | 0.073 | 1.286 | 0.441 | 0.690 | 0.793 | 0.670 |
| Creatinine, µmol/L | 0.765 | 0.924 | 0.071 | 1.286 | 0.770 | 0.693 | 0.117 | 0.659 |
| Liver function/hepatic damage | ||||||||
| AST, U/L | 0.883 | 0.925 | 0.973 | 1.311 | 0.720 | 0.691 | 0.989 | 0.670 |
| GGT, U/L | 0.801 | 0.924 | 0.532 | 1.306 | 0.130 | 0.678 | 0.573 | 0.666 |
| BILt, µmol/L | 0.080 | 0.914 | 0.280 | 1.299 | 0.584 | 0.691 | 0.053 | 0.660 |
| Albumin, g/L | 0.099 | 0.910 | 0.360 | 1.302 | 0.627 | 0.690 | 0.119 | 0.654 |
| ALP, U/L | 0.337 | 0.919 | 0.267 | 1.297 | 0.015 | 0.674 | <0.001 | 0.618 |
| PON, U/mL | 0.942 | 0.926 | 0.824 | 1.309 | 0.920 | 0.693 | 0.776 | 0.669 |
| Oxidative stress metabolites | ||||||||
| ROMt, mgH2O2/100 mL | 0.243 | 0.924 | 0.126 | 1.291 | 0.052 | 0.673 | 0.597 | 0.668 |
| AOPP, µmol/L | 0.014 | 0.896 | 0.017 | 1.266 | 0.177 | 0.677 | 0.080 | 0.659 |
| FRAP, µmol/L | 0.642 | 0.922 | 0.178 | 1.292 | 0.172 | 0.685 | 0.498 | 0.668 |
| SHp, µmol/L | 0.241 | 0.914 | 0.459 | 1.306 | 0.044 | 0.683 | 0.663 | 0.667 |
| Inflammation/innate immunity | ||||||||
| Ceruloplasmin, µmol/L | 0.010 | 0.913 | 0.030 | 1.280 | 0.012 | 0.664 | 0.611 | 0.668 |
| PROTt, g/L | 0.578 | 0.923 | 0.785 | 1.308 | 0.464 | 0.688 | 0.481 | 0.666 |
| Globulins, g/L | 0.748 | 0.926 | 0.361 | 1.300 | 0.962 | 0.693 | 0.354 | 0.662 |
| Haptoglobin, g/L | 0.873 | 0.925 | 0.338 | 1.296 | 0.060 | 0.680 | 0.626 | 0.667 |
| MPO, U/L | 0.320 | 0.916 | 0.126 | 1.284 | 0.895 | 0.693 | 0.498 | 0.667 |
| Minerals | ||||||||
| Ca, mmol/L | 0.432 | 0.919 | 0.702 | 1.307 | 0.785 | 0.691 | 0.381 | 0.665 |
| P, mmol/L | 0.122 | 0.910 | 0.333 | 1.305 | 0.690 | 0.692 | 0.844 | 0.668 |
| Mg, mmol/L | 0.302 | 0.928 | 0.361 | 1.305 | 0.450 | 0.690 | 0.541 | 0.668 |
| Na, mmol/L | 0.872 | 0.925 | 0.473 | 1.302 | 0.954 | 0.693 | 0.782 | 0.669 |
| K, mmol/L | 0.149 | 0.923 | 0.271 | 1.303 | 0.482 | 0.690 | 0.663 | 0.668 |
| Cl, mmol/L | 0.871 | 0.925 | 0.618 | 1.313 | 0.168 | 0.684 | 0.131 | 0.661 |
| Zn, µmol/L | 0.396 | 0.922 | 0.109 | 1.288 | 0.430 | 0.689 | 0.206 | 0.668 |
| Traits 1 | F5-Production | F6-SCFA 2 | F7-OCFA 3 | F8-RBH 4 | ||||
|---|---|---|---|---|---|---|---|---|
| p-Value | RMSE 5 | p-Value | RMSE 5 | p-Value | RMSE 2 | p-Value | RMSE 5 | |
| Individual sources of variation 6 | ||||||||
| DIM, classes 7 | 0.139 | 1.868 | 0.381 | 2.900 | 0.569 | 1.159 | 0.701 | 0.922 |
| Parity, classes 8 | 0.013 | 0.689 | 0.839 | 0.468 | ||||
| Body measure | ||||||||
| BCS, classes | 0.850 | 1.783 | 0.744 | 2.848 | 0.195 | 1.162 | 0.057 | 0.920 |
| Ultrasound measurements | ||||||||
| pTAG, mg/g | 0.030 | 1.834 | 0.044 | 2.829 | 0.344 | 1.159 | 0.143 | 0.919 |
| PVA, mm2 | 0.783 | 1.878 | 0.151 | 2.855 | 0.052 | 1.164 | 0.126 | 0.939 |
| PVD, mm | 0.665 | 1.880 | 0.350 | 2.890 | 0.227 | 1.167 | 0.738 | 0.938 |
| LD, mm | 0.929 | 1.918 | 0.895 | 2.938 | 0.815 | 1.166 | 0.521 | 0.949 |
| Hematochemical parameters | ||||||||
| Hematocrit, L/L | 0.569 | 1.978 | 0.386 | 2.954 | 0.220 | 1.184 | 0.695 | 0.955 |
| Energy-related metabolites | ||||||||
| Glucose, mmol/L | 0.670 | 1.957 | 0.678 | 2.938 | 0.039 | 1.159 | 0.016 | 0.921 |
| Cholesterol, mmol/L | 0.004 | 1.877 | 0.082 | 2.901 | 0.955 | 1.189 | 0.016 | 0.926 |
| NEFA, mmol/L | 0.146 | 1.908 | 0.403 | 2.924 | 0.352 | 1.177 | 0.899 | 0.954 |
| BHB, mmol/L | 0.457 | 1.970 | 0.613 | 2.956 | 0.003 | 1.135 | 0.253 | 0.948 |
| Urea, mmol/L | 0.777 | 1.966 | 0.700 | 2.954 | 0.108 | 1.167 | 0.673 | 0.953 |
| Creatinine, µmol/L | 0.103 | 1.950 | 0.292 | 2.941 | 0.886 | 1.189 | 0.384 | 0.947 |
| Liver function/hepatic damage | ||||||||
| AST, U/L | 0.754 | 1.976 | 0.972 | 2.965 | 0.678 | 1.185 | 0.689 | 0.951 |
| GGT, U/L | 0.480 | 1.961 | 0.177 | 2.913 | 0.046 | 1.162 | 0.145 | 0.944 |
| BILt, µmol/L | 0.576 | 1.970 | 0.148 | 2.928 | 0.155 | 1.168 | 0.853 | 0.950 |
| Albumin, g/L | 0.037 | 1.896 | 0.137 | 2.880 | <0.001 | 1.066 | 0.011 | 0.909 |
| ALP, U/L | 0.863 | 1.964 | 0.798 | 2.962 | 0.105 | 1.171 | 0.232 | 0.945 |
| PON, U/mL | 0.168 | 1.953 | 0.379 | 2.944 | 0.187 | 1.177 | 0.752 | 0.951 |
| Oxidative stress metabolites | ||||||||
| ROMt, mgH2O2/100 mL | 0.724 | 1.965 | 0.455 | 2.950 | 0.527 | 1.177 | 0.387 | 0.948 |
| AOPP, µmol/L | 0.170 | 1.904 | 0.262 | 2.899 | 0.367 | 1.178 | 0.015 | 0.955 |
| FRAP, µmol/L | 0.618 | 1.934 | 0.597 | 2.909 | 0.991 | 1.189 | 0.912 | 0.957 |
| SHp, µmol/L | 0.378 | 1.958 | 0.400 | 2.947 | 0.032 | 1.159 | 0.504 | 0.944 |
| Inflammation/innate immunity | ||||||||
| Ceruloplasmin, µmol/L | 0.677 | 1.962 | 0.686 | 2.958 | 0.016 | 1.147 | 0.526 | 0.950 |
| PROTt, g/L | 0.705 | 1.968 | 0.800 | 2.962 | 0.023 | 1.157 | 0.395 | 0.949 |
| Globulins, g/L | 0.396 | 1.966 | 0.276 | 2.940 | 0.021 | 1.159 | 0.727 | 0.948 |
| Haptoglobin, g/L | 0.082 | 1.937 | 0.171 | 2.931 | 0.647 | 1.184 | 0.675 | 0.953 |
| MPO, U/L | 0.411 | 1.968 | 0.722 | 2.960 | 0.898 | 1.189 | 0.836 | 0.952 |
| Minerals | ||||||||
| Ca, mmol/L | 0.537 | 1.953 | 0.771 | 2.959 | 0.648 | 1.188 | 0.882 | 0.952 |
| P, mmol/L | 0.748 | 1.976 | 0.347 | 2.945 | 0.902 | 1.190 | 0.592 | 0.952 |
| Mg, mmol/L | 0.786 | 1.966 | 0.717 | 2.959 | 0.111 | 1.161 | 0.645 | 0.948 |
| Na, mmol/L | 0.596 | 1.957 | 0.654 | 2.955 | 0.510 | 1.184 | 0.143 | 0.947 |
| K, mmol/L | 0.263 | 1.962 | 0.072 | 2.914 | 0.742 | 1.187 | 0.495 | 0.949 |
| Cl, mmol/L | 0.441 | 1.969 | 0.171 | 2.931 | 0.915 | 1.188 | 0.765 | 0.951 |
| Zn, µmol/L | 0.737 | 1.971 | 0.171 | 2.931 | 0.972 | 1.189 | 0.090 | 0.949 |
| FA Factors (F) 1 | Indicators of Metabolic Stress 2 | Physiological Meaning |
|---|---|---|
| F1-de novo/ preformed | AOPP, µmol/L (↓) CP, µmol/L (↓) | Enzymatic activities decreased during inflammatory and oxidative stress |
| F2-BCFA | AOPP, µmol/L (↑) CP, µmol/L (↑) pTAG, mg/g (↓) Cholesterol, µmol/L (↑) | Negatively related with negative energetic balance and reduced feed intake Involvement in oxidative pathways Reported proinflammatory activities |
| F3-LC-PUFA | BCS, classes (↑) CP, µmol/L (↑) | Mirror of energy balance and lipid accumulation in the organism Positively associated with marker of inflammation and hepatic damage |
| F4-alternative RBH ALA | pTAG, mg/g (↓) NEFA, mmol/L (↓) Glucose, µmol/L (↑) ALP, U/L (↑) | Positively associated with marker of inflammation and hepatic damage Inverse relationship with negative energy balance and hepatic lipid accumulation |
| F5-production | pTAG, mg/g (↓) Cholesterol, mmol/L (↑) Albumin, U/L (↑) | Inverse relationship with negative energy balance and hepatic lipid accumulation |
| F6-SCFA | pTAG, mg/g (↓) | Dependent from glucose metabolism Inverse relationship with negative energy balance and hepatic lipid accumulation |
| F7-OCFA | GGT, U/L (↑) SHp, µmol/L (↑) Albumin, U/L (↑) BHB, mmol/L (↓) CP, µmol/L (↓) Globulins, U/L (↓) | Positively associated with antioxidant and anti-inflammatory activity Increased OCFA levels during hepatic damage Negatively associated to ketonemia, might be used as indicator of negative energy balance |
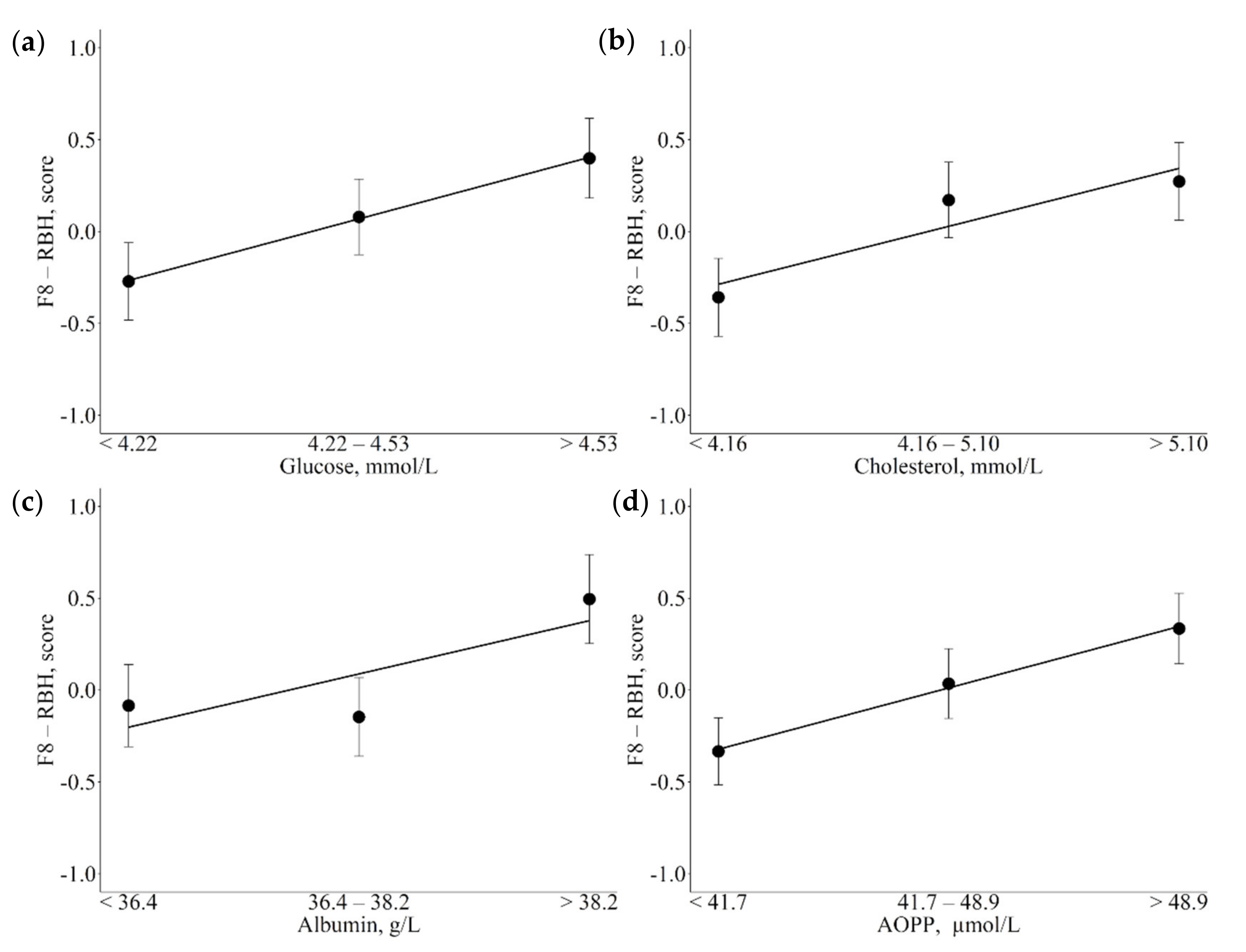
| F8-RBH | Glucose, µmol/L (↑) Cholesterol, mmol/L (↑) Albumin, U/L (↑) AOPP, µmol/L (↑) | Negatively related with negative energetic balance and reduced feed intake that reduced rumen enzymatic activities Enhanced by glucose availability Involvement in oxidative pathways |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannuzzi, D.; Toscano, A.; Pegolo, S.; Gallo, L.; Tagliapietra, F.; Mele, M.; Minuti, A.; Trevisi, E.; Ajmone Marsan, P.; Schiavon, S.; et al. Associations between Milk Fatty Acid Profile and Body Condition Score, Ultrasound Hepatic Measurements and Blood Metabolites in Holstein Cows. Animals 2022, 12, 1202. https://doi.org/10.3390/ani12091202
Giannuzzi D, Toscano A, Pegolo S, Gallo L, Tagliapietra F, Mele M, Minuti A, Trevisi E, Ajmone Marsan P, Schiavon S, et al. Associations between Milk Fatty Acid Profile and Body Condition Score, Ultrasound Hepatic Measurements and Blood Metabolites in Holstein Cows. Animals. 2022; 12(9):1202. https://doi.org/10.3390/ani12091202
Chicago/Turabian StyleGiannuzzi, Diana, Alessandro Toscano, Sara Pegolo, Luigi Gallo, Franco Tagliapietra, Marcello Mele, Andrea Minuti, Erminio Trevisi, Paolo Ajmone Marsan, Stefano Schiavon, and et al. 2022. "Associations between Milk Fatty Acid Profile and Body Condition Score, Ultrasound Hepatic Measurements and Blood Metabolites in Holstein Cows" Animals 12, no. 9: 1202. https://doi.org/10.3390/ani12091202








