Can Grassland Chemical Quality Be Quantified Using Transform Near-Infrared Spectroscopy?
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Forage Sample Set
- i.
- Old alfalfa meadows re-colonized by spontaneous species (n = 35);
- ii.
- Grass–legume mixtures recently established (n = 30);
- iii.
- Old legume mixtures grassland re-colonized by native species (n = 60);
- iv.
- Alfalfa crops recently established (n = 25).
2.2. Sample Preparation and Spectral Measurement
2.3. Chemical Analysis
2.4. Statistical Analysis
3. Results
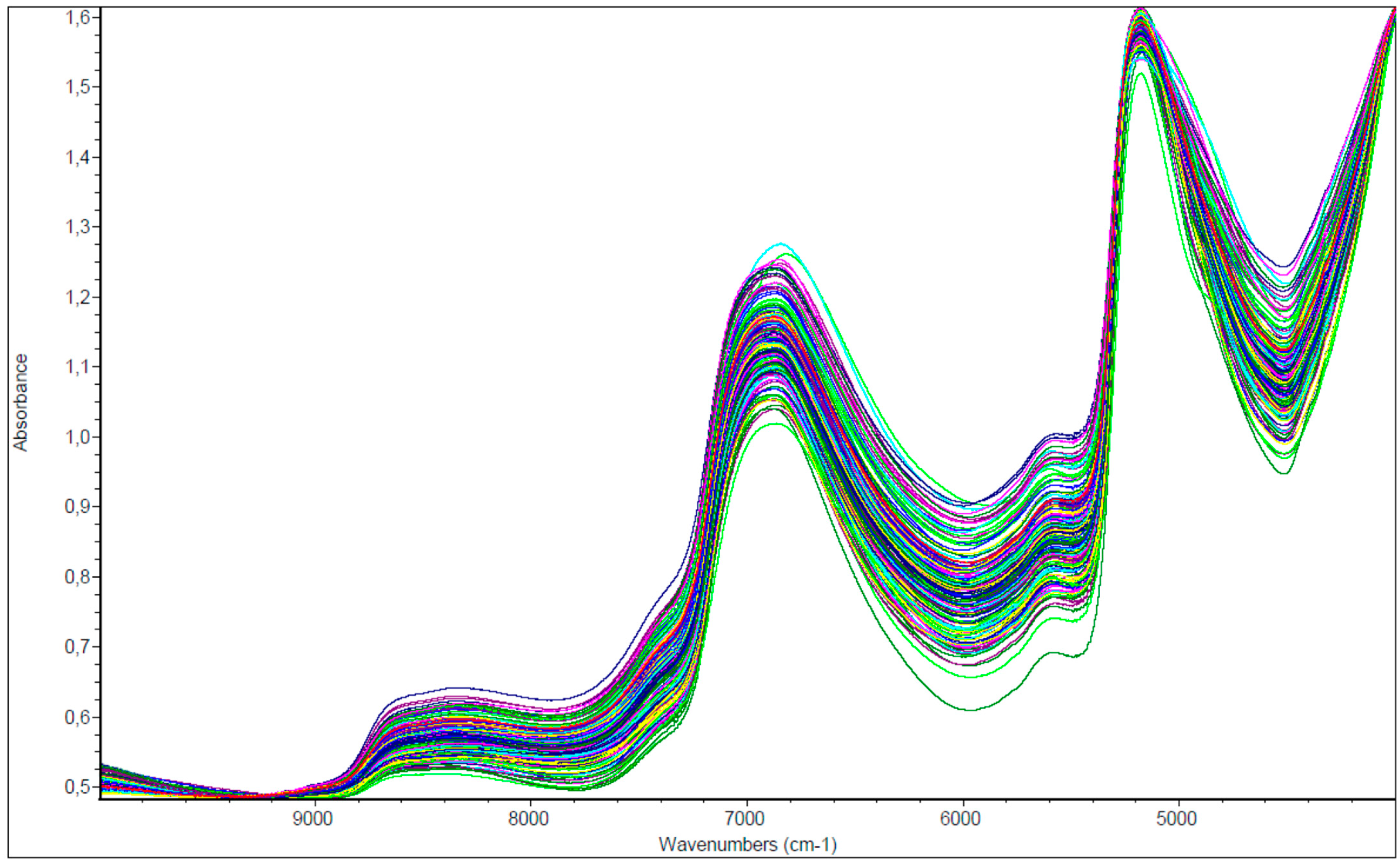
3.1. Near-Infrared Spectra
3.2. Descriptive Statistics
3.3. NIRS Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Argenti, G.; Bottai, L.; Chiesi, M.; Maselli, F.; Staglianò, N.; Targetti, S. Analysis and assessment of mountain pastures by integration of multispectral and ancillary data. Ital. J. Remote Sens. 2011, 43, 45. [Google Scholar] [CrossRef]
- Blüthgen, N.; Simons, N.K.; Jung, K.; Prati, D.; Renner, S.C.; Boch, S.; Fischer, M.; Hölzel, N.; Klaus, V.H.; Kleinebecker, T.; et al. Land use imperils plant and animal community stability through changes in asynchrony rather than diversity. Nat. Commun. 2016, 7, 10697. [Google Scholar] [CrossRef] [Green Version]
- Giustini, L.; Acciaioli, A.; Argenti, G. Apparent balance of nitrogen and phosphorus in dairy farms in Mugello (Italy). Ital. J. Anim. Sci. 2007, 6, 175–185. [Google Scholar] [CrossRef]
- Gossner, M.M.; Lewinsohn, T.M.; Kahl, T.; Grassein, F.; Boch, S.; Prati, D.; Birkhofer, K.; Renner, S.C.; Sikorski, J.; Wubet, T.; et al. Land-use intensification causes multitrophic homogenization of grassland communities. Nature 2016, 540, 266–269. [Google Scholar] [CrossRef]
- Targetti, S.; Messeri, A.; Staglianò, N.; Argenti, G. Leaf functional traits for the assessment of succession following management in semi-natural grasslands: A case study in the North Apennines, Italy. Appl. Veg. Sci. 2013, 16, 325–332. [Google Scholar] [CrossRef]
- Pittarello, M.; Probo, M.; Perotti, E.; Lonati, M.; Lombardi, G.; Ravetto Enri, S. Grazing management plans improve pasture selection by cattle and forage quality in sub-Alpine and Alpine grasslands. J. Mt. Sci. 2019, 16, 2126–2135. [Google Scholar] [CrossRef]
- Berauer, B.J.; Wilfahrt, P.A.; Reu, B.; Schuchardt, M.A.; Garcia-Franco, N.; Zistl-Schlingmann, M.; Dannenmann, M.; Kiese, R.; Kühnel, A.; Jentsch, A. Predicting forage quality of species-rich pasture grasslands using vis-NIRS to reveal effects of management intensity and climate change. Agric. Ecosyst. Environ. 2020, 296, 106929. [Google Scholar] [CrossRef]
- Movedi, E.; Bellocchi, G.; Argenti, G.; Paleari, L.; Vesely, F.; Staglianò, N.; Dibari, C.; Confalonieri, R. Development of generic crop models for simulation of multi-species plant communities in Mown grasslands. Ecol. Model. 2019, 401, 111–128. [Google Scholar] [CrossRef]
- Khalsa, J.; Fricke, T.; Weisser, W.W.; Weigelt, A.; Wachendorf, M. Effects of functional groups and species richness on biomass constituents relevant for combustion: Results from a grassland diversity experiment. Grass Forage Sci. 2012, 67, 569–588. [Google Scholar] [CrossRef]
- Boob, M.; Elsaesser, M.; Thumm, U.; Hartung, J.; Lewandowski, I. Harvest time determines quality and usability of biomass from Lowland Hay Meadows. Agriculture 2019, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- Niu, K.; He, J.; Zhang, S.; Lechowicz, M.J. Tradeoffs between forage quality and soil fertility: Lessons from Himalayan Rangelands. Agric. Ecosyst. Environ. 2016, 234, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Argenti, G.; Cervasio, F.; Ponzetta, M.P. Control of bracken (Pteridium aquilinum) and feeding preferences in pastures grazed by wild ungulates in an area of the northern apennines (Italy). Ital. J. Anim. Sci. 2012, 11, e62. [Google Scholar] [CrossRef] [Green Version]
- Pierik, M.E.; Gusmeroli, F.; Marianna, G.D.; Tamburini, A.; Bocchi, S. Meadows species composition, biodiversity and forage value in an alpine district: Relationships with environmental and dairy farm management variables. Agric. Ecosyst. Environ. 2017, 244, 14–21. [Google Scholar] [CrossRef]
- Dibari, C.; Argenti, G.; Catolfi, F.; Moriondo, M.; Staglianò, N.; Bindi, M. Pastoral suitability driven by future climate change along the Apennines. Ital. J. Agron. 2015, 10, 659. [Google Scholar] [CrossRef] [Green Version]
- Bokobza, L. Near infrared spectroscopy. J. Near Infrared Spectrosc. 1998, 6, 3–17. [Google Scholar] [CrossRef]
- Roberts, C.A.; Workman, S.; Reeves, J.B. Near Infrared Spectroscopy in Agriculture; American Society of Agronomy Inc.: Madison, WI, USA, 2003. [Google Scholar]
- Ozaki, Y. Near-infrared spectroscopy—Its versatility in analytical chemistry. Anal. Sci. 2012, 28, 545–563. [Google Scholar] [CrossRef] [Green Version]
- Ibáñez, L.; Alomar, D. Prediction of the chemical composition and fermentation parameters of pasture silage by near infrered reflectance spectroscopy (NIRS). Chil. J. Agric. Res. 2008, 68, 352–359. [Google Scholar] [CrossRef]
- Restaino, E.A.; Fernández, E.G.; Manna, A.L.; Cozzolino, D. Prediction of the nutritive value of pasture silage by near infrared spectroscopy (NIRS). Chil. J. Agric. Res. 2009, 69, 560–566. [Google Scholar] [CrossRef]
- Halgerson, J.L.; Sheaffer, C.C.; Martin, N.P.; Peterson, P.R.; Weston, S.J. Near-infrared reflectance spectroscopy prediction of leaf and mineral concentrations in Alfalfa. Agron. J. 2004, 96, 344–351. [Google Scholar]
- Brogna, N.; Pacchioli, M.T.; Immovilli, A.; Ruozzi, F.; Ward, R.; Formigoni, A. The use of near-infrared reflectance spectroscopy (NIRS) in the prediction of chemical composition and in vitro neutral detergent fiber (NDF) digestibility of Italian Alfalfa Hay. Ital. J. Anim. Sci. 2009, 8, 271–273. [Google Scholar] [CrossRef] [Green Version]
- Hetta, M.; Mussadiq, Z.; Wallsten, J.; Halling, M.; Swensson, C.; Geladi, P. Prediction of nutritive values, morphology and agronomic characteristics in forage maize using two applications of NIRS spectrometry. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2017, 67, 326–333. [Google Scholar] [CrossRef]
- Stubbs, T.L.; Kennedy, A.C.; Fortuna, A.-M. Using NIRS to predict fiber and nutrient content of dryland cereal cultivars. J. Agric. Food Chem. 2010, 58, 398–403. [Google Scholar] [CrossRef]
- Olsoy, P.J.; Griggs, T.C.; Ulappa, A.C.; Gehlken, K.; Shipley, L.A.; Shewmaker, G.E.; Forbey, J.S. Nutritional analysis of sagebrush by near-infrared reflectance spectroscopy. J. Arid. Environ. 2016, 134, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Andrés, S.; Giráldez, F.J.; López, S.; Mantecón, Á.R.; Calleja, A. Nutritive evaluation of herbage from permanent meadows by near-infrared reflectance spectroscopy: 1. Prediction of chemical composition and in vitro digestibility. J. Sci. Food Agric. 2005, 85, 1564–1571. [Google Scholar] [CrossRef]
- Ronchi, B.; Bernabucci, U.; Carlini, P.; Danieli, P.P. Quality evaluation of regional forage resources by means of near infrared reflectance spectroscopy. Ital. J. Anim. Sci. 2010, 3, 363–376. [Google Scholar] [CrossRef]
- Lobos, I.; Gou, P.; Hube, S.; Saldaña, R.; Alfaro, M. Evaluation of potential nirs to predict pastures nutritive value. J. Soil Sci. Plant Nutr. 2013, 13, 463–468. [Google Scholar] [CrossRef] [Green Version]
- Parrini, S.; Acciaioli, A.; Crovetti, A.; Bozzi, R. Use of FT-NIRS for determination of chemical components and nutritional value of natural pasture. Ital. J. Anim. Sci. 2018, 17, 87–91. [Google Scholar] [CrossRef] [Green Version]
- Alomar, D.; Fuchslocher, R.; Cuevas, J.; Mardones, R.; Cuevas, E. Prediction of the composition of fresh pastures by near Infrared reflectance or interactance-reflectance spectroscopy. Chil. J. Agric. Res. 2009, 69, 198–206. [Google Scholar] [CrossRef] [Green Version]
- Reddersen, B.; Fricke, T.; Wachendorf, M. Effects of sample preparation and measurement standardization on the NIRS calibration quality of nitrogen, ash and NDFom content in extensive experimental grassland biomass. Anim. Feed. Sci. Technol. 2013, 183, 77–85. [Google Scholar] [CrossRef]
- Anderson, T.M.; Griffith, D.M.; Grace, J.B.; Lind, E.M.; Adler, P.B.; Biederman, L.A.; Blumenthal, D.M.; Daleo, P.; Firn, J.; Hagenah, N.; et al. Herbivory and eutrophication mediate grassland plant nutrient responses across a global climatic gradient. Ecology 2018, 99, 822–831. [Google Scholar] [CrossRef] [Green Version]
- Locher, F.; Heuwinkel, H.; Gutser, R.; Schmidhalter, U. The legume content in multispecies mixtures as estimated with near infrared reflectance spectroscopy method validation. Agron. J. 2005, 97, 18–25. [Google Scholar] [CrossRef]
- Karayilanli, E.; Cherney, J.H.; Sirois, P.; Kubinec, D.; Cherney, D.J.R. Botanical composition prediction of alfalfa–grass mixtures using NIRS: Developing a robust calibration. Crop. Sci. 2016, 56, 3361–3366. [Google Scholar] [CrossRef]
- Wilson, R.L.; Bionaz, M.; MacAdam, J.W.; Beauchemin, K.A.; Naumann, H.D.; Ates, S. Milk production, nitrogen utilization, and methane emissions of dairy cows grazing grass, forb, and legume-based pastures. J. Anim. Sci. 2020, 98, skaa220. [Google Scholar] [CrossRef]
- Horwitz, W.; Chichilo, P.; Reynolds, H. Official Methods of Analysis of the Association of Official Analytical Chemists; Association of Official Analytical Chemists: Washington, DC, USA, 1970. [Google Scholar]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Leonardi, L.; Burns, D.H. Quantitative Measurements in Scattering Media: Photon Time-of-Flight Analysis with Analytical Descriptors. Available online: https://journals.sagepub.com/doi/abs/10.1366/0003702991947270 (accessed on 22 October 2021).
- Thermo Fischer Scientific TQ Analyst Software: User Guide. Madison, WI, USA. 2011. Available online: https://www.google.com.hk/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&ved=2ahUKEwjxlJnj_oz1AhUIZt4KHU8iD5gQFnoECAcQAQ&url=https%3A%2F%2Fassets.thermofisher.com%2FTFS-Assets%2FCAD%2FSpecification-Sheets%2FD14232~.pdf&usg=AOvVaw103i3h6zGKsnJZBJxuYroL (accessed on 22 October 2021).
- Williams, P.; Dardenne, P.; Flinn, P. Tutorial: Items to be included in a report on a near infrared spectroscopy project. J. Near Infrared Spectrosc. 2017, 25, 85–90. [Google Scholar] [CrossRef]
- Perez-Marin, D.C.; Garrido-Varo, A.; Guerrero-Ginel, J.E.; Gomez-Cabrera, A. Near-infrared reflectance spectroscopy (NIRS) for the mandatory labelling of compound feedingstuffs: Chemical composition and open-declaration. Anim. Feed Sci. Technol. 2004, 116, 333–349. [Google Scholar] [CrossRef]
- Williams, P. The RPD statistic: A tutorial note. NIR News 2014, 25, 22–26. [Google Scholar] [CrossRef]
- Williams, P.C.; Sobering, D.C. Comparison of commercial near infrared transmittance and reflectance instruments for analysis of whole grains and seeds. J. Near Infrared Spectrosc. JNIRS 1993, 1, 25–32. [Google Scholar] [CrossRef]
- Williams, P.C.; Norris, K. Near-Infrared Technology in the Agricultural and Food Industries; American Association of Cereals Chemists: St. Paul, MN, USA, 2001. [Google Scholar]
- Barbin, D.F.; Kaminishikawahara, C.M.; Soares, A.L.; Mizubuti, I.Y.; Grespan, M.; Shimokomaki, M.; Hirooka, E.Y. Prediction of chicken quality attributes by near infrared spectroscopy. Food Chem. 2015, 168, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Millmier, A.; Lorimor, J.C.; Hurburgh, C.R., Jr.; Fulhage, C.; Hattey, J. Near-infrared sensing of manure ingredients. Trans. ASAE 2000, 43, 903–908. [Google Scholar] [CrossRef]
- Magwaza, L.S.; Opara, U.L.; Nieuwoudt, H.; Cronje, P.J.R.; Saeys, W.; Nicolaï, B. NIR spectroscopy applications for internal and external quality analysis of citrus fruit—A review. Food Bioprocess Technol. 2012, 5, 425–444. [Google Scholar] [CrossRef]
- Manley, M. Near-infrared spectroscopy and hyperspectral imaging: Non-destructive analysis of biological materials. Chem. Soc. Rev. 2014, 43, 8200–8214. [Google Scholar] [CrossRef] [Green Version]
- Stuart, B.H. Infrared Spectroscopy: Fundamentals and Applications; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2004. [Google Scholar]
- Lugassi, R.; Chudnovsky, A.; Zaady, E.; Dvash, L.; Goldshleger, N. Estimating pasture quality of fresh vegetation based on spectral slope of mixed data of dry and fresh vegetation—Method development. Remote Sens. 2015, 7, 8045–8066. [Google Scholar] [CrossRef] [Green Version]
- Kokaly, R.F. Investigating a physical basis for spectroscopic estimates of leaf nitrogen concentration. Remote Sens. Environ. 2001, 75, 153–161. [Google Scholar] [CrossRef]
- Schwanninger, M.; Rodrigues, J.C.; Fackler, K. A review of band assignments in near infrared spectra of wood and wood components. J. Near Infrared Spectrosc. JNIRS 2011, 19, 287–308. [Google Scholar] [CrossRef]
- Li, X.; Sun, C.; Zhou, B.; He, Y. Determination of hemicellulose, cellulose and lignin in moso bamboo by near infrared spectroscopy. Sci. Rep. 2015, 5, 17210. [Google Scholar] [CrossRef]
- Barton, F.E.; Himmelsbach, D.S. Two-dimensional vibrational spectroscopy II: Correlation of the absorptions of lignins in the mid- and near-infrared. Appl. Spectrosc. 1993, 47, 1920–1925. [Google Scholar] [CrossRef]
- Osborne, B.G. Near-infrared spectroscopy in food analysis. In Encyclopedia of Analytical Chemistry; American Cancer Society: Washington, DC, USA, 2006; ISBN 9780470027318. [Google Scholar] [CrossRef]
- Zude, M.; Pflanz, M.; Kaprielian, C.; Aivazian, B.L. NIRS as a tool for precision horticulture in the citrus industry. Biosyst. Eng. 2008, 99, 455–459. [Google Scholar] [CrossRef]
- Cougnon, M.; Waes, C.V.; Dardenne, P.; Baert, J.; Reheul, D. Comparison of near infrared reflectance spectroscopy calibration strategies for the botanical composition of grass-clover mixtures. Grass Forage Sci. 2014, 69, 167–175. [Google Scholar] [CrossRef]
- Biewer, S.; Fricke, T.; Wachendorf, M. Development of canopy reflectance models to predict forage quality of legume–grass mixtures. Crop. Sci. 2009, 49, 1917–1926. [Google Scholar] [CrossRef]
- Elle, O.; Richter, R.; Vohland, M.; Weigelt, A. Fine root lignin content is well predictable with near-infrared spectroscopy. Sci. Rep. 2019, 9, 6396. [Google Scholar] [CrossRef]
- Pasquini, C. Near infrared spectroscopy: Fundamentals, practical aspects and analytical applications. J. Braz. Chem. Soc. 2003, 14, 198–219. [Google Scholar] [CrossRef] [Green Version]
- Rijal, D.; Walsh, K.B.; Subedi, P.P.; Ashwath, N. Quality estimation of agave tequilana leaf for bioethanol production. J. Near Infrared Spectrosc. 2016, 24, 453–465. [Google Scholar] [CrossRef] [Green Version]
- Rinnan, Å.; van den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Yang, Z.; Nie, G.; Pan, L.; Zhang, Y.; Huang, L.; Ma, X.; Zhang, X. Development and validation of near-infrared spectroscopy for the prediction of forage quality parameters in lolium multiflorum. PeerJ 2017, 5, e3867. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Zhu, R.; Xu, R.; Zhang, W.; Shen, Y.; Zhang, Y. Evaluation of leymus chinensis quality using near-infrared reflectance spectroscopy with three different statistical analyses. PeerJ 2015, 3, e1416. [Google Scholar] [CrossRef] [Green Version]
- Fekadu, D.; Bediye, S.; Kehaliw, A.; Daba, T.; Kitaw, G.; Assefa, G. Near infrared reflectance spectroscopy (NIRS) for determination of chemical entities of natural pasture from Ethiopia. Agric. Biol. J. N. Am. 2010, 1, 919–922. [Google Scholar] [CrossRef]
- Norman, H.C.; Hulm, E.; Humphries, A.W.; Hughes, S.J.; Vercoe, P.E.; Norman, H.C.; Hulm, E.; Humphries, A.W.; Hughes, S.J.; Vercoe, P.E. Broad near-infrared spectroscopy calibrations can predict the nutritional value of >100 forage species within the Australian feedbase. Anim. Prod. Sci. 2020, 60, 1111–1122. [Google Scholar] [CrossRef]
| Parameters | Calibration | Validation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean | Median | SD | min | max | n | Mean | Median | SD | min | max | |
| Dry matter g/100 g | 115 | 20.04 | 18.50 | 6.71 | 11.14 | 43.12 | 30 | 20.29 | 18.87 | 5.25 | 12.22 | 32.00 |
| Crude protein | 118 | 17.73 | 18.25 | 4.74 | 7.43 | 25.79 | 30 | 17.69 | 18.64 | 4.54 | 10.04 | 25.37 |
| Ash | 114 | 10.52 | 10.61 | 2.05 | 4.49 | 15.33 | 30 | 10.86 | 10.96 | 1.23 | 7.70 | 12.71 |
| Crude fat | 115 | 2.28 | 2.32 | 0.40 | 1.25 | 3.09 | 30 | 2.35 | 2.34 | 0.34 | 1.49 | 2.87 |
| NDF | 116 | 51.11 | 50.61 | 8.59 | 32.77 | 71.68 | 30 | 48.53 | 48.08 | 7.07 | 40.04 | 65.83 |
| ADF | 117 | 34.85 | 34.96 | 6.94 | 22.13 | 45.49 | 30 | 35.61 | 36.06 | 4.94 | 25.00 | 43.00 |
| ADL | 116 | 6.65 | 6.94 | 2.40 | 1.72 | 12.36 | 30 | 7.27 | 7.55 | 1.39 | 4.00 | 9.25 |
| Parameters | FPLS | Range WN (cm−1) | Math Treat. | Calibration | Validation | RPD | RER | ||
|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSEC | R2v | RMSEv | ||||||
| Dry matter g/100 g | 6 | 5118–6817 4800–7100 | 1; 3; 5 | 0.951 | 1.88 | 0.938 | 1.97 | 2.7 | 10.1 |
| Crude protein | 6 | 4800–5200 6200–7200 | 2; 3; 5 | 0.905 | 1.45 | 0.901 | 1.48 | 3.1 | 10.4 |
| Ash | 3 | 4000–9000 | 2; 4; 5 | 0.837 | 0.73 | 0.754 | 1.01 | 1.2 | 5.0 |
| Crude fat | 3 | 5100–9200 | 1; 3; 5 | 0.737 | 0.66 | 0.652 | 0.24 | 1.4 | 5.8 |
| NDF | 5 | 5500–6200 | 2; 3; 6 | 0.911 | 2.45 | 0.885 | 2.47 | 2.9 | 10.4 |
| ADF | 5 | 5500–6200 | 1; 4; 6 | 0.946 | 1.06 | 0.936 | 1.23 | 4.0 | 14.6 |
| ADL | 10 | 5183–8333 | 1; 4; 6 | 0.908 | 0.63 | 0.880 | 0.60 | 2.3 | 8.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parrini, S.; Staglianò, N.; Bozzi, R.; Argenti, G. Can Grassland Chemical Quality Be Quantified Using Transform Near-Infrared Spectroscopy? Animals 2022, 12, 86. https://doi.org/10.3390/ani12010086
Parrini S, Staglianò N, Bozzi R, Argenti G. Can Grassland Chemical Quality Be Quantified Using Transform Near-Infrared Spectroscopy? Animals. 2022; 12(1):86. https://doi.org/10.3390/ani12010086
Chicago/Turabian StyleParrini, Silvia, Nicolina Staglianò, Riccardo Bozzi, and Giovanni Argenti. 2022. "Can Grassland Chemical Quality Be Quantified Using Transform Near-Infrared Spectroscopy?" Animals 12, no. 1: 86. https://doi.org/10.3390/ani12010086






