SINE Insertion in the Intron of Pig GHR May Decrease Its Expression by Acting as a Repressor
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Acquisition and Structural Variation Prediction for GH/IGF Axis Genes
2.2. Retrotransposon Annotation and Insertion Polymorphic Prediction
2.3. RIP Verification and Genotyping
2.4. Dual-Luciferase Reporter Assay
2.5. Expression Analysis
2.6. Statistical Analysis
3. Results
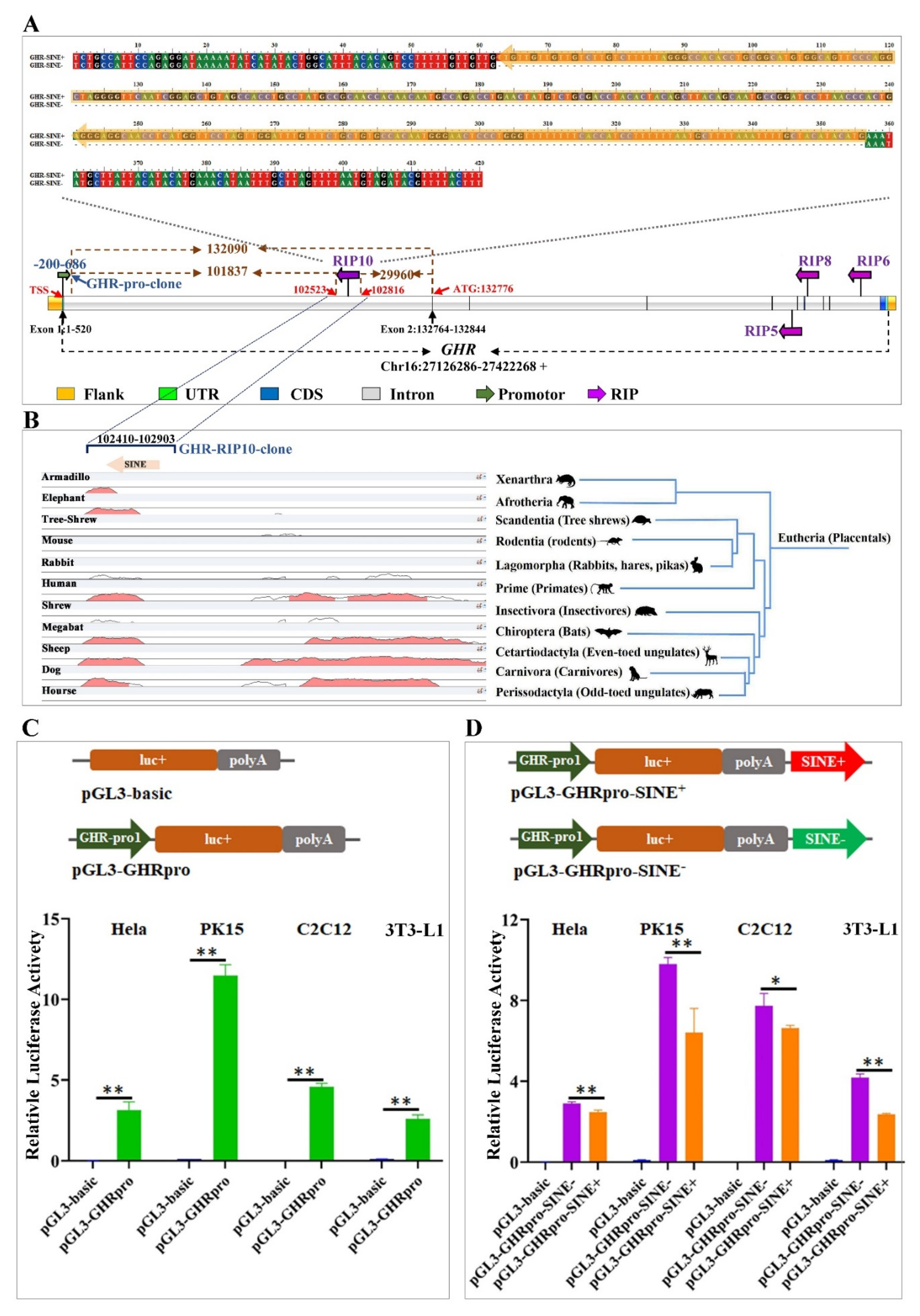
3.1. Five RIPs Generated by Retrotransposon Insertions in the Pig GH/IGF Axis Genes
3.2. RIP Distribution in Different Pig Breeds
3.3. SINE Insertion in the First Intron of GHR May Repress the Promoter Activity
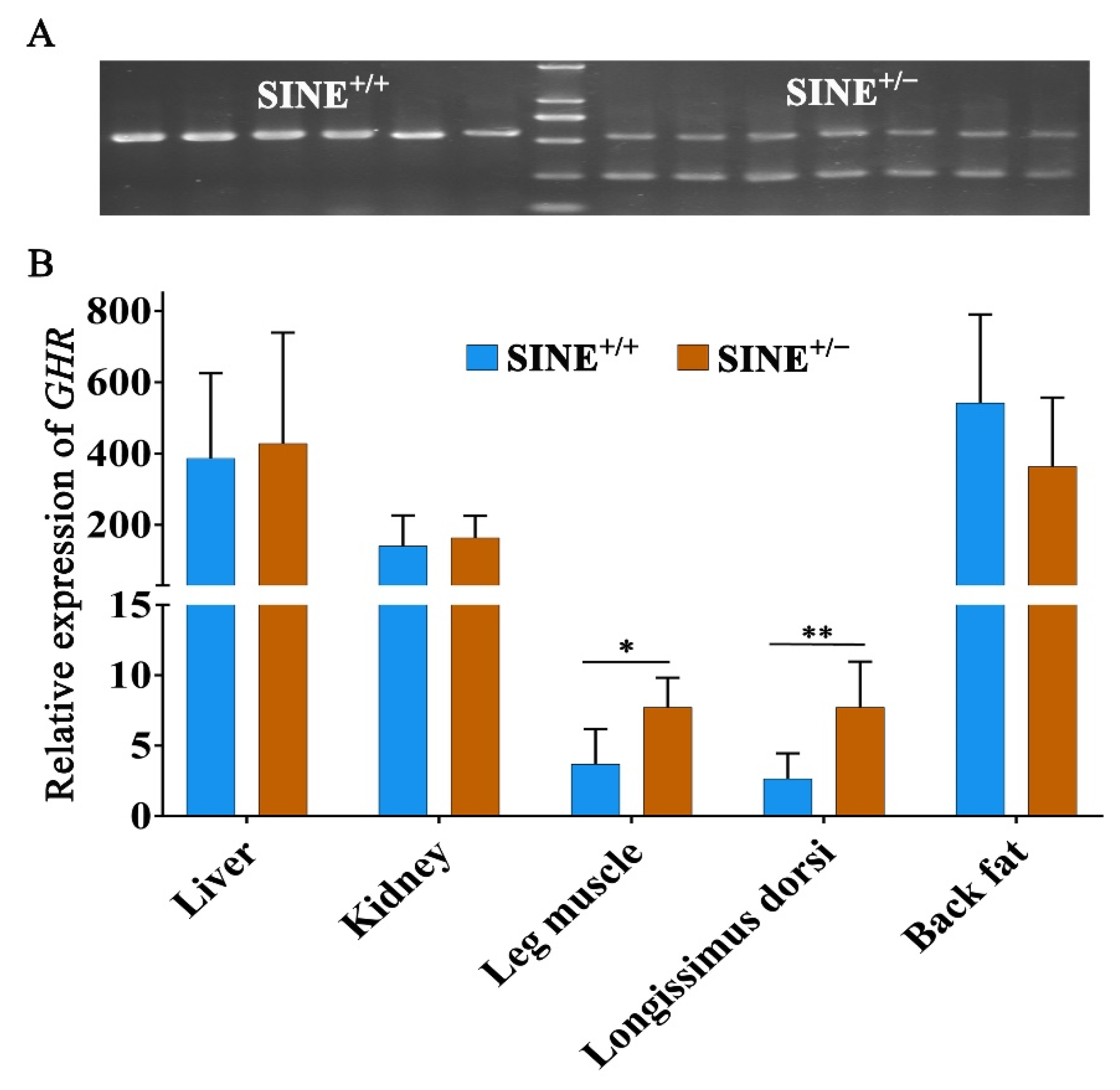
3.4. Decreased Expression of GHR in Muscle Associated with the SINE Insertion in the First Intron
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mathews, L.S.; Norstedt, G.; Palmiter, R.D. Regulation of insulin-like growth factor I gene expression by growth hormone. Proc. Natl. Acad. Sci. USA 1986, 83, 9343–9347. [Google Scholar] [CrossRef] [PubMed]
- Dixit, M.; Poudel, S.B.; Yakar, S. Effects of GH/IGF axis on bone and cartilage. Mol. Cell. Endocrinol. 2021, 519, 111052. [Google Scholar] [CrossRef] [PubMed]
- Holmes, S.J.; Shalet, S.M. Role of Growth Hormone and Sex Steroids in Achieving and Maintaining Normal Bone Mass. Horm. Res. 1996, 45, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Sanyal, S.; Chattopadhyay, N. The role of estrogen in bone growth and formation: Changes at puberty. Cell Health Cytoskelet. 2010, 3, 2–12. [Google Scholar] [CrossRef][Green Version]
- Lupu, F.; Terwilliger, J.D.; Lee, K.; Segre, G.V.; Efstratiadis, A. Roles of growth hormone and insulin-like growth factor 1 in mouse postnatal growth. Dev. Biol. 2001, 229, 141–162. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, R.G.; Rosenbloom, A.L.; Guevara-aguirre, J. Growth Hormone (GH) Insensitivity Due to Primary GH Receptor Deficiency. Endocr. Rev. 1994, 15, 369–390. [Google Scholar] [CrossRef]
- Domené, H.M.; Fierro-Carrión, G. Genetic disorders of GH action pathway. Growth Horm. IGF Res. 2018, 38, 19–23. [Google Scholar] [CrossRef]
- Stevenson, A.E.; Evans, B.A.J.; Gevers, E.F.; Elford, C.; McLeod, R.W.J.; Perry, M.J.; El-Kasti, M.M.; Coschigano, K.T.; Kopchick, J.J.; Evans, S.L.; et al. Does adiposity status influence femoral cortical strength in rodent models of growth hormone deficiency? Am. J. Physiol. Metab. 2009, 296, E147–E156. [Google Scholar] [CrossRef]
- O’Neill, B.T.; Lauritzen, H.P.M.M.; Hirshman, M.F.; Smyth, G.; Goodyear, L.J.; Kahn, C.R. Differential Role of Insulin/IGF-1 Receptor Signaling in Muscle Growth and Glucose Homeostasis. Cell Rep. 2015, 11, 1220–1235. [Google Scholar] [CrossRef]
- Boucher, J.; Softic, S.; El Ouaamari, A.; Krumpoch, M.T.; Kleinridders, A.; Kulkarni, R.N.; O’Neill, B.T.; Kahn, C.R. Differential Roles of Insulin and IGF-1 Receptors in Adipose Tissue Development and Function. Diabetes 2016, 65, 2201–2213. [Google Scholar] [CrossRef]
- Farabaugh, S.M.; Boone, D.N.; Lee, A.V. Role of IGF1R in Breast Cancer Subtypes, Stemness, and Lineage Differentiation. Front. Endocrinol. 2015, 6, 59. [Google Scholar] [CrossRef]
- Mazzoccoli, G.; Giuliani, F.; Inglese, M.; Marzulli, N.; Dagostino, M.P.; De Cata, A.; Greco, A.; Carughi, S.; Tarquini, R. Chronobiologic study of the GH-IGF1 axis and the ageing immune system. J. Appl. Biomed. 2010, 8, 213–226. [Google Scholar] [CrossRef]
- Colao, A.; Marzullo, P.; Di Somma, C.; Lombardi, G. Growth hormone and the heart. Clin. Endocrinol. 2001, 54, 137–154. [Google Scholar] [CrossRef]
- Juul, A.; Scheike, T.; Davidsen, M.; Gyllenborg, J.; Jørgensen, T. Low serum insulin-like growth factor I is associated with increased risk of ischemic heart disease: A population-based case-control study. Circulation 2002, 106, 939–944. [Google Scholar] [CrossRef]
- Bondanelli, M.; Ambrosio, M.R.; Onofri, A.; Bergonzoni, A.; Lavezzi, S.; Zatelli, M.C.; Valle, D.; Basaglia, N.; degli Uberti, E.C. Predictive Value of Circulating Insulin-Like Growth Factor I Levels in Ischemic Stroke Outcome. J. Clin. Endocrinol. Metab. 2006, 91, 3928–3934. [Google Scholar] [CrossRef] [PubMed]
- Platt, R.N.; Vandewege, M.W.; Ray, D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosom. Res. 2018, 26, 25–43. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Chen, C.; Wang, W.; Wang, X.; Shen, D.; Wang, S.; Wang, Y.; Gao, B.; Wimmers, K.; Mao, J.; Li, K.; et al. Retrotransposons evolution and impact on lncRNA and protein coding genes in pigs. Mob. DNA 2019, 10, 19. [Google Scholar] [CrossRef]
- Garcia-Perez, J.L.; Widmann, T.J.; Adams, I.R. The impact of transposable elements on mammalian development. Development 2016, 143, 4101–4114. [Google Scholar] [CrossRef]
- Arkhipova, I.R.; Yushenova, I.A. Giant Transposons in Eukaryotes: Is Bigger Better? Genome Biol. Evol. 2019, 11, 906–918. [Google Scholar] [CrossRef]
- Kaaij, L.J.T.; Mohn, F.; van der Weide, R.H.; de Wit, E.; Bühler, M. The ChAHP Complex Counteracts Chromatin Looping at CTCF Sites that Emerged from SINE Expansions in Mouse. Cell 2019, 178, 1437–1451. [Google Scholar] [CrossRef]
- Göke, J.; Ng, H.H. CTRL + INSERT: Retrotransposons and their contribution to regulation and innovation of the transcriptome. EMBO Rep. 2016, 17, 1131–1144. [Google Scholar] [CrossRef]
- Feschotte, C. The contribution of transposable elements ot the evolution of regulatory networks. Nat. Rev. Genet. 2008, 9, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Hancks, D.C.; Kazazian, H.H. Roles for retrotransposon insertions in human disease. Mob. DNA 2016, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ran, X.; Niu, X.; Li, S.; Wang, J.; Zhang, Q. Insertion of 275-bp SINE into first intron of PDIA4 gene is associated with litter size in Xiang pigs. Anim. Reprod. Sci. 2018, 195, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Chen, C.; Chen, W.; Wang, X.; Wang, W.; Gao, B.; Wimmers, K.; Mao, J.; Song, C. Two new SINE insertion polymorphisms in pig Vertnin (VRTN) gene revealed by comparative genomic alignment. J. Integr. Agric. 2020, 19, 2514–2522. [Google Scholar] [CrossRef]
- Mikawa, S.; Sato, S.; Nii, M.; Morozumi, T.; Yoshioka, G.; Imaeda, N.; Yamaguchi, T.; Hayashi, T.; Awata, T. Identification of a second gene associated with variation in vertebral number in domestic pigs. BMC Genet. 2011, 12, 5. [Google Scholar] [CrossRef]
- Fontanesi, L.; Scotti, E.; Buttazzoni, L.; Dall’Olio, S.; Russo, V. Investigation of a Short Interspersed Nuclear Element Polymorphic Site in the Porcine Vertnin Gene: Allele Frequencies and Association Study with Meat Quality, Carcass and Production Traits in Italian Large White pigs. Ital. J. Anim. Sci. 2014, 13, 3090. [Google Scholar] [CrossRef]
- Gray, M.M.; Sutter, N.B.; Ostrander, E.A.; Wayne, R.K. The IGF1small dog haplotype is derived from Middle Eastern grey wolves. BMC Biol. 2010, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Qu, L.; Yao, J.; Yang, X.; Li, G.; Zhang, Y.; Li, J.; Wang, X.; Bai, J.; Xu, G.; et al. An EAV-HP Insertion in 5′ Flanking Region of SLCO1B3 Causes Blue Eggshell in the Chicken. PLoS Genet. 2013, 9, e1003183. [Google Scholar] [CrossRef] [PubMed]
- Giuffra, E.; Törnsten, A.; Marklund, S.; Bongcam-Rudloff, E.; Chardon, P.; Kijas, J.M.H.; Anderson, S.I.; Archibald, A.L.; Andersson, L. A large duplication associated with dominant white color in pigs originated by homologous recombination between LINE elements flanking KIT. Mamm. Genome 2002, 13, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Sironen, A.; Uimari, P.; Iso-Touru, T.; Vilkki, J. L1 insertion within SPEF2 gene is associated with increased litter size in the Finnish Yorkshire population. J. Anim. Breed. Genet. 2012, 129, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.G.; Yue, M.; Gu, Y.; Gu, W.W.; Wang, Y.J. Single-nucleotide polymorphism analysis of GH, GHR, and IGF-1 genes in minipigs. Brazilian J. Med. Biol. Res. 2014, 47, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Dettori, M.L.; Pazzola, M.; Paschino, P.; Amills, M.; Vacca, G.M. Association between the GHR, GHRHR, and IGF1 gene polymorphisms and milk yield and quality traits in Sarda sheep. J. Dairy Sci. 2018, 101, 9978–9986. [Google Scholar] [CrossRef]
- Kawashima, C.; Munakata, M.; Matsui, M.; Miyamoto, A.; Kida, K.; Shimizu, T. Polymorphism in promoter region of growth hormone receptor is associated with potential production capacity of insulin-like growth factor-1 in pre-pubertal Holstein heifers. J. Anim. Physiol. Anim. Nutr. 2016, 100, 1037–1040. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Ahmed, I.; Liu, L.; Liu, Y.; Xu, Z.; Duan, X.; Li, Q.; Dou, T.; Gu, D.; Rong, H.; et al. Selection for growth rate and body size have altered the expression profiles of somatotropic axis genes in chickens. PLoS ONE 2018, 13, e0195378. [Google Scholar] [CrossRef]
- Sutter, N.B.; Bustamante, C.D.; Chase, K.; Gray, M.M.; Zhao, K.; Zhu, L.; Padhukasahasram, B.; Karlins, E.; Davis, S.; Jones, P.G.; et al. A single IGF1 allele is a major determinant of small size in dogs. Science 2007, 316, 112–115. [Google Scholar] [CrossRef]
- Hoopes, B.C.; Rimbault, M.; Liebers, D.; Ostrander, E.A.; Sutter, N.B. The insulin-like growth factor 1 receptor (IGF1R) contributes to reduced size in dogs. Mamm. Genome 2012, 23, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Nishimura, D. RepeatMasker. Biotech Softw. Internet Rep. 2000, 1, 36–39. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.C.; Boyle, T.B.J.; Ye, Z.H.; Mao, J.X. POPGENE, the user-friendly shareware for population genetic analysis. Mol. Biol. Biotechnol. Centre Univ. Alberta Canada 1997, 10, 295–301. [Google Scholar]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Min, X.; Gui An, H.; Feng Yan, M.; Juan, L.; Ya jun, W. Characterization of Growth Hormone Receptor (GHR) Gene Promoter in pigs: Molecular Cloning and Functional Analysis. J. Sichuan Univ. 2015, 52, 1353–1358. [Google Scholar]
- Zhang, Z.D.; Paccanaro, A.; Fu, Y.; Weissman, S.; Weng, Z.; Chang, J.; Snyder, M.; Gerstein, M.B. Statistical analysis of the genomic distribution and correlation of regulatory elements in the ENCODE regions. Genome Res. 2007, 17, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Michael, T.P. Plant genome size variation: Bloating and purging DNA. Brief. Funct. Genomics 2014, 13, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Shen, D.; Xue, S.; Chen, C.; Cui, H.; Song, C. The contribution of transposable elements to size variations between four teleost genomes. Mob. DNA 2016, 7, 4. [Google Scholar] [CrossRef]
- Chalopin, D.; Naville, M.; Plard, F.; Galiana, D.; Volff, J.-N. Comparative Analysis of Transposable Elements Highlights Mobilome Diversity and Evolution in Vertebrates. Genome Biol. Evol. 2015, 7, 567–580. [Google Scholar] [CrossRef]
- Kumar, A.; Hirochika, H. Applications of retrotransposons as genetic tools in plant biology. Trends Plant Sci. 2001, 6, 127–134. [Google Scholar] [CrossRef]
- Kalendar, R.; Flavell, A.J.; Ellis, T.H.N.; Sjakste, T.; Moisy, C.; Schulman, A.H. Analysis of plant diversity with retrotransposon-based molecular markers. Heredity 2011, 106, 520–530. [Google Scholar] [CrossRef]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Kazazian, H.H.; Moran, J.V. The impact of L1 retrotransposons on the human genome. Nat. Genet. 1998, 19, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Goodier, J.L. Restricting retrotransposons: A review. Mob. DNA 2016, 7, 16. [Google Scholar] [CrossRef]
- Chen, C.; D’Alessandro, E.; Murani, E.; Zheng, Y.; Giosa, D.; Yang, N.; Wang, X.; Gao, B.; Li, K.; Wimmers, K.; et al. SINE jumping contributes to large-scale polymorphisms in the pig genomes. Mob. DNA 2021. [Google Scholar] [CrossRef]
- Chen, C.; Wang, X.; Zong, W.; D’Alessandro, E.; Giosa, D.; Guo, Y.; Mao, J.; Song, C. Genetic Diversity and Population Structures in Chinese Miniature Pigs Revealed by SINE Retrotransposon Insertion Polymorphisms, a New Type of Genetic Markers. Animals 2021, 11, 1136. [Google Scholar] [CrossRef]
- Almeida, L.M.; Silva, I.T.; Silva, W.A., Jr.; Castro, J.P.; Riggs, P.K.; Carareto, C.M.; Amaral, M.E.J. The contribution of transposable elements to Bos taurus gene structure. Gene 2007, 390, 180–189. [Google Scholar] [CrossRef]
- Burns, K.H.; Boeke, J.D. Human Transposon Tectonics. Cell 2012, 149, 740–752. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; Sela, N.; Ast, G. TranspoGene and microTranspoGene: Transposed elements influence on the transcriptome of seven vertebrates and invertebrates. Nucleic Acids Res. 2008, 36, D47–D52. [Google Scholar] [CrossRef][Green Version]
- Abdollahi Mandoulakani, B.; Piri, Y.; Darvishzadeh, R.; Bernoosi, I.; Jafari, M. Retroelement Insertional Polymorphism and Genetic Diversity in Medicago sativa Populations Revealed by IRAP and REMAP Markers. Plant Mol. Biol. Report. 2012, 30, 286–296. [Google Scholar] [CrossRef]
- Allen, T.A.; Von Kaenel, S.; Goodrich, J.A.; Kugel, J.F. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004, 11, 816–821. [Google Scholar] [CrossRef]
- Mariner, P.D.; Walters, R.D.; Espinoza, C.A.; Drullinger, L.F.; Wagner, S.D.; Kugel, J.F.; Goodrich, J.A. Human Alu RNA Is a Modular Transacting Repressor of mRNA Transcription during Heat Shock. Mol. Cell 2008, 29, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Ponicsan, S.L.; Kugel, J.F.; Goodrich, J.A. Genomic gems: SINE RNAs regulate mRNA production. Curr. Opin. Genet. Dev. 2010, 20, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, C.A.; Allen, T.A.; Hieb, A.R.; Kugel, J.F.; Goodrich, J.A. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 2004, 11, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Anamika; Chakraborty, D.; Kumar, D.; Azhar, P.M.; Singh, S.G.; Singh, S.; Sudan, A. Growth Hormone Receptor (GHR) gene and its applicationsin livestock: A review. Agric. Rev. 2016, 37, 250–254. [Google Scholar]

| No. of Large SVs | |||||
|---|---|---|---|---|---|
| Total | GHR | GH | IGF1 | IGF1R | |
| SVs ≥ 51 bp | 92 | 47 | 3 | 7 | 35 |
| Predicted RIPs | 42 | 26 | 0 | 3 | 13 |
| Confirmed RIPs | 5 | 4 | 0 | 1 | 0 |
| RIP Name | Mutation Type | Chr | Begin | End | TE Type | Orientation Relative to Gene | Length | Gene Structure |
|---|---|---|---|---|---|---|---|---|
| GHR-RIP10 | Deletion | Chr16 | 27228809 | 27229102 | SINEA6 | Antisense | 294 | Intron 1 |
| GHR-RIP5 | Deletion | Chr16 | 27388037 | 27388337 | SINEA1 | Antisense | 301 | Intron 5 |
| GHR-RIP8 | Deletion | Chr16 | 27393631 | 27393854 | L1D20 | Antisense | 318 | Intron 7 |
| GHR-RIP6 | Deletion | Chr16 | 27412697 | 27412983 | SINEA1 | Antisense | 287 | Intron 9 |
| IGF1-RIP2 | Deletion | Chr5 | 81790669 | 81790888 | SINEA1 | Sense | 300 | Intron 3 |
| RIP Name | Polymorphic Breeds | Population Size | Genotype Frequency | Allele Frequency | Hardy-Weinberg Equilibrium Test/p Value | PIC | |||
|---|---|---|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | + | − | |||||
| GHR-RIP5 | Wuzhishan | 32 | 0.79 | 0.15 | 0.06 | 0.86 | 0.14 | 0.0455 | 0.2118 |
| Landrace | 32 | 0.00 | 1.00 | 0.00 | 0.50 | 0.50 | 0.0000 | 0.3750 | |
| Large White | 32 | 0.84 | 0.16 | 0.00 | 0.92 | 0.08 | 0.6317 | 0.1364 | |
| Sujiang | 32 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 | NA | 0.0000 | |
| Sushan | 32 | 0.00 | 1.00 | 0.00 | 0.50 | 0.50 | 0.0000 | 0.3750 | |
| Bama | 21 | 0.57 | 0.43 | 0.00 | 0.79 | 0.21 | 0.2114 | 0.2768 | |
| Mingguang small ear | 20 | 0.75 | 0.15 | 0.10 | 0.83 | 0.17 | 0.0269 | 0.2424 | |
| GHR-RIP6 | Wuzhishan | 32 | 0.00 | 1.00 | 0.00 | 0.50 | 0.50 | 0.0000 | 0.3750 |
| Landrace | 32 | 0.00 | 0.81 | 0.19 | 0.40 | 0.60 | 0.0001 | 0.3648 | |
| Large White | 32 | 0.00 | 0.94 | 0.06 | 0.47 | 0.53 | 0.0000 | 0.3741 | |
| Sujiang | 32 | 0.00 | 1.00 | 0.00 | 0.50 | 0.50 | 0.0000 | 0.3750 | |
| Sushan | 32 | 0.00 | 1.00 | 0.00 | 0.50 | 0.50 | 0.0000 | 0.3750 | |
| Bama | 21 | 0.00 | 1.00 | 0.00 | 0.50 | 0.50 | 0.0000 | 0.3750 | |
| Mingguang small ear | 20 | 0.00 | 0.95 | 0.05 | 0.48 | 0.52 | 0.0001 | 0.3746 | |
| GHR-RIP8 | Wuzhishan | 32 | 0.56 | 0.41 | 0.03 | 0.77 | 0.23 | 0.4553 | 0.2915 |
| Landrace | 32 | 0.03 | 0.41 | 0.56 | 0.23 | 0.77 | 0.4553 | 0.2915 | |
| Large White | 32 | 0.28 | 0.53 | 0.19 | 0.55 | 0.45 | 0.6841 | 0.3725 | |
| Sujiang | 32 | 0.81 | 0.19 | 0.00 | 0.91 | 0.09 | 0.5584 | 0.1504 | |
| Sushan | 32 | 0.19 | 0.75 | 0.06 | 0.56 | 0.44 | 0.0030 | 0.3714 | |
| Bama | 21 | 0.95 | 0.05 | 0.00 | 0.98 | 0.02 | 0.9110 | 0.0384 | |
| Mingguang small ear | 20 | 0.85 | 0.15 | 0.00 | 0.92 | 0.08 | 0.3362 | 0.1364 | |
| GHR-RIP10 | Wuzhishan | 32 | 0.19 | 0.44 | 0.37 | 0.41 | 0.59 | 0.5984 | 0.3668 |
| Landrace | 32 | 0.00 | 0.19 | 0.81 | 0.09 | 0.91 | 0.0000 | 0.1504 | |
| Large White | 32 | 0.34 | 0.53 | 0.13 | 0.61 | 0.39 | 0.5121 | 0.3626 | |
| Sujiang | 32 | 0.47 | 0.47 | 0.06 | 0.70 | 0.30 | 0.4872 | 0.3318 | |
| Sushan | 32 | 0.44 | 0.44 | 0.12 | 0.66 | 0.34 | 0.3917 | 0.3481 | |
| Bama | 21 | 0.00 | 0.76 | 0.24 | 0.38 | 0.62 | 0.0048 | 0.3602 | |
| Mingguang small ear | 20 | 0.65 | 0.35 | 0.00 | 0.83 | 0.17 | 0.3428 | 0.2424 | |
| IGF1-RIP2 | Wuzhishan | 32 | 0.13 | 0.53 | 0.34 | 0.39 | 0.61 | 0.5121 | 0.3626 |
| Landrace | 32 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 | NA | 0.0000 | |
| Large White | 32 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 | NA | 0.0000 | |
| Sujiang | 32 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 | NA | 0.0000 | |
| Sushan | 32 | 0.00 | 1.00 | 0.00 | 0.50 | 0.50 | 0.0000 | 0.3750 | |
| Bama | 21 | 0.48 | 0.43 | 0.09 | 0.69 | 0.31 | 0.9903 | 0.3363 | |
| Mingguang small ear | 20 | 0.75 | 0.25 | 0.00 | 0.88 | 0.12 | 0.5229 | 0.1889 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Zheng, Y.; Wang, M.; Murani, E.; D’Alessandro, E.; Moawad, A.S.; Wang, X.; Wimmers, K.; Song, C. SINE Insertion in the Intron of Pig GHR May Decrease Its Expression by Acting as a Repressor. Animals 2021, 11, 1871. https://doi.org/10.3390/ani11071871
Chen C, Zheng Y, Wang M, Murani E, D’Alessandro E, Moawad AS, Wang X, Wimmers K, Song C. SINE Insertion in the Intron of Pig GHR May Decrease Its Expression by Acting as a Repressor. Animals. 2021; 11(7):1871. https://doi.org/10.3390/ani11071871
Chicago/Turabian StyleChen, Cai, Yao Zheng, Mengli Wang, Eduard Murani, Enrico D’Alessandro, Ali Shoaib Moawad, Xiaoyan Wang, Klaus Wimmers, and Chengyi Song. 2021. "SINE Insertion in the Intron of Pig GHR May Decrease Its Expression by Acting as a Repressor" Animals 11, no. 7: 1871. https://doi.org/10.3390/ani11071871
APA StyleChen, C., Zheng, Y., Wang, M., Murani, E., D’Alessandro, E., Moawad, A. S., Wang, X., Wimmers, K., & Song, C. (2021). SINE Insertion in the Intron of Pig GHR May Decrease Its Expression by Acting as a Repressor. Animals, 11(7), 1871. https://doi.org/10.3390/ani11071871









