miR-F4-C12 Functions on the Regulation of Adipose Accumulation by Targeting PIK3R1 in Castrated Male Pigs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Cell Differentiation and Transfection
2.3. Oil Red O Staining
2.4. RNA Preparation and qRT-PCR
2.5. Plasmid Construction
2.6. Dual-Luciferase Reporter Assay
2.7. Protein Isolation and Western Blot Analysis
2.8. Statistical Data Analysis
3. Result
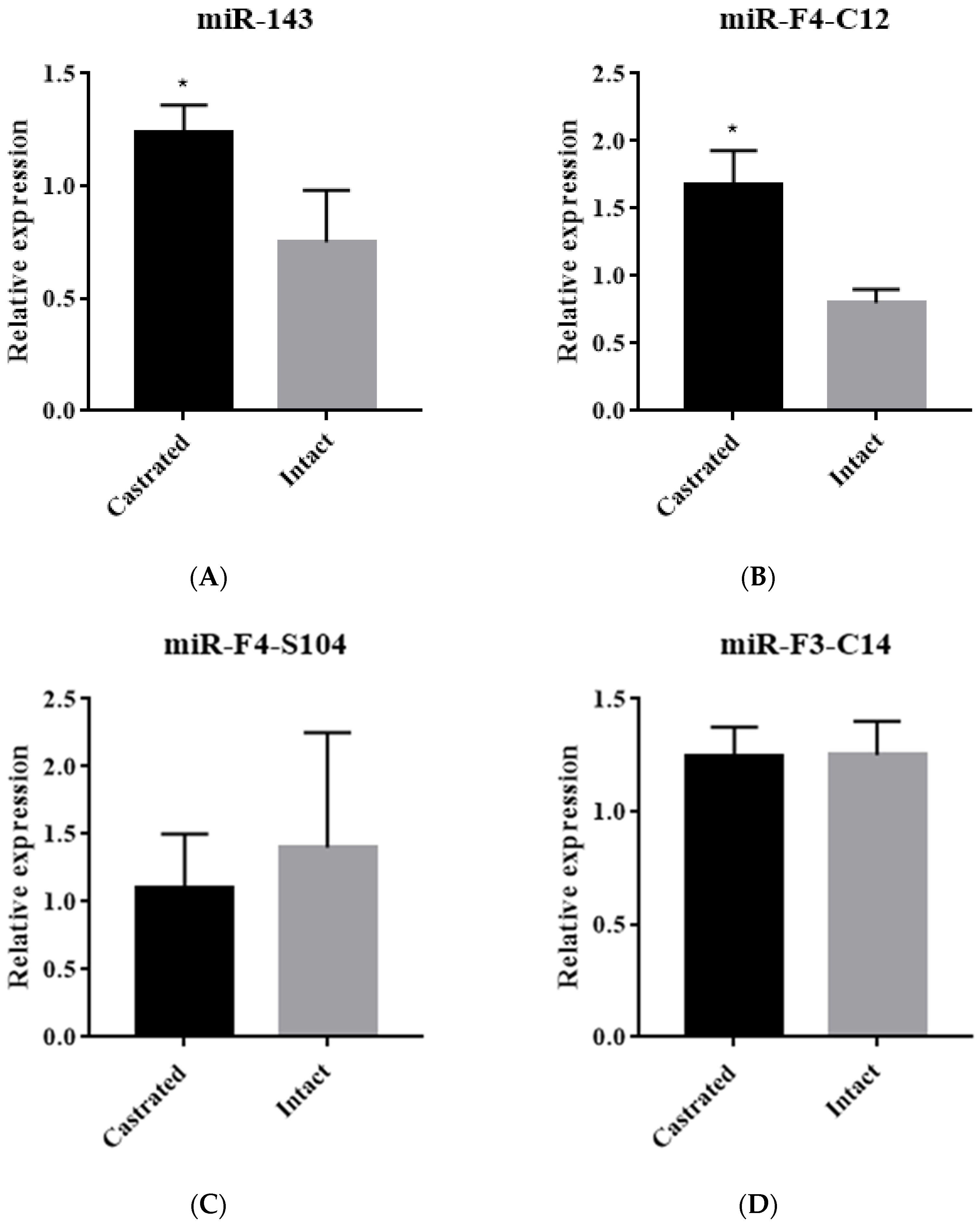
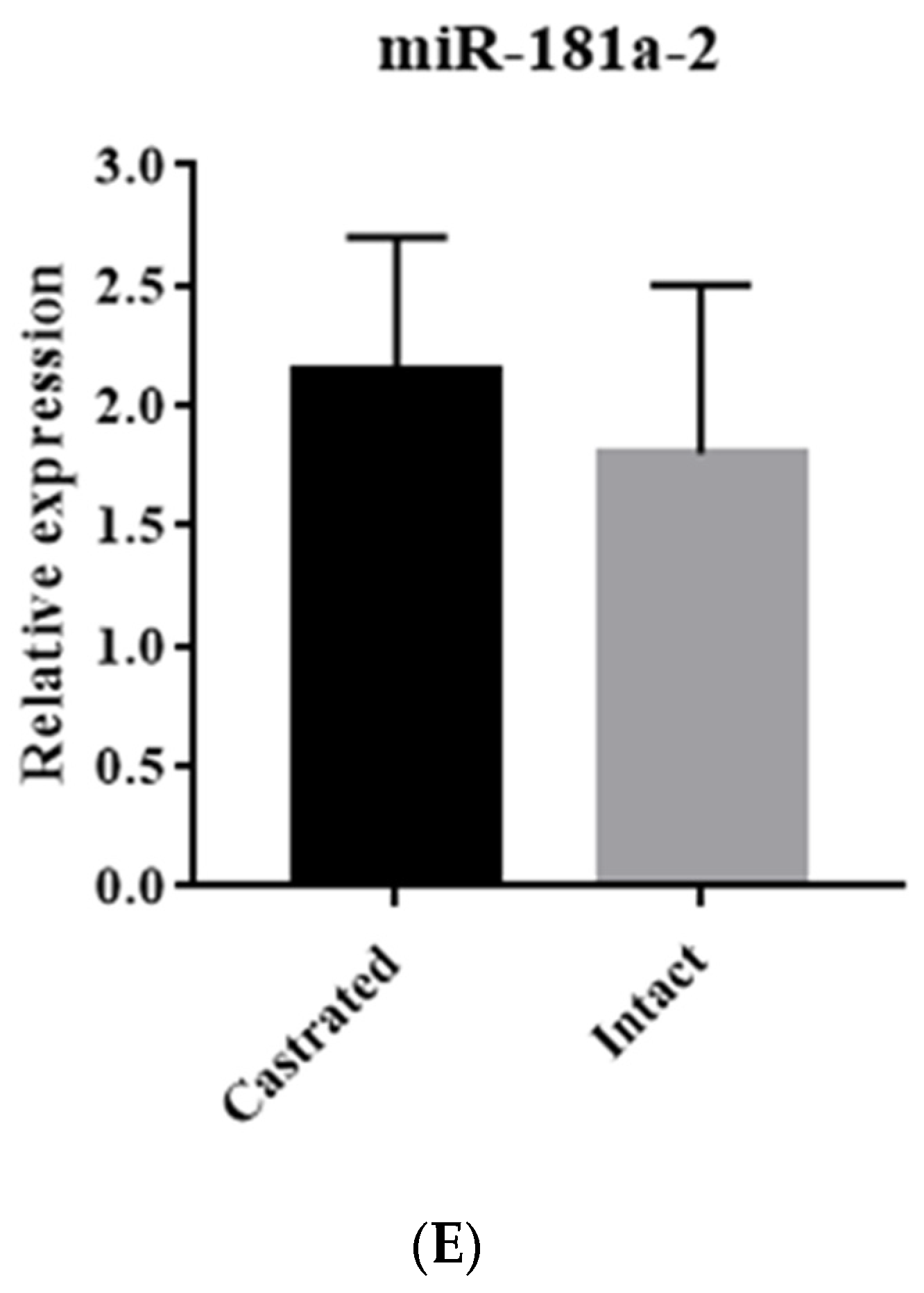
3.1. Identification of miRNA Expression in Backfat Tissue
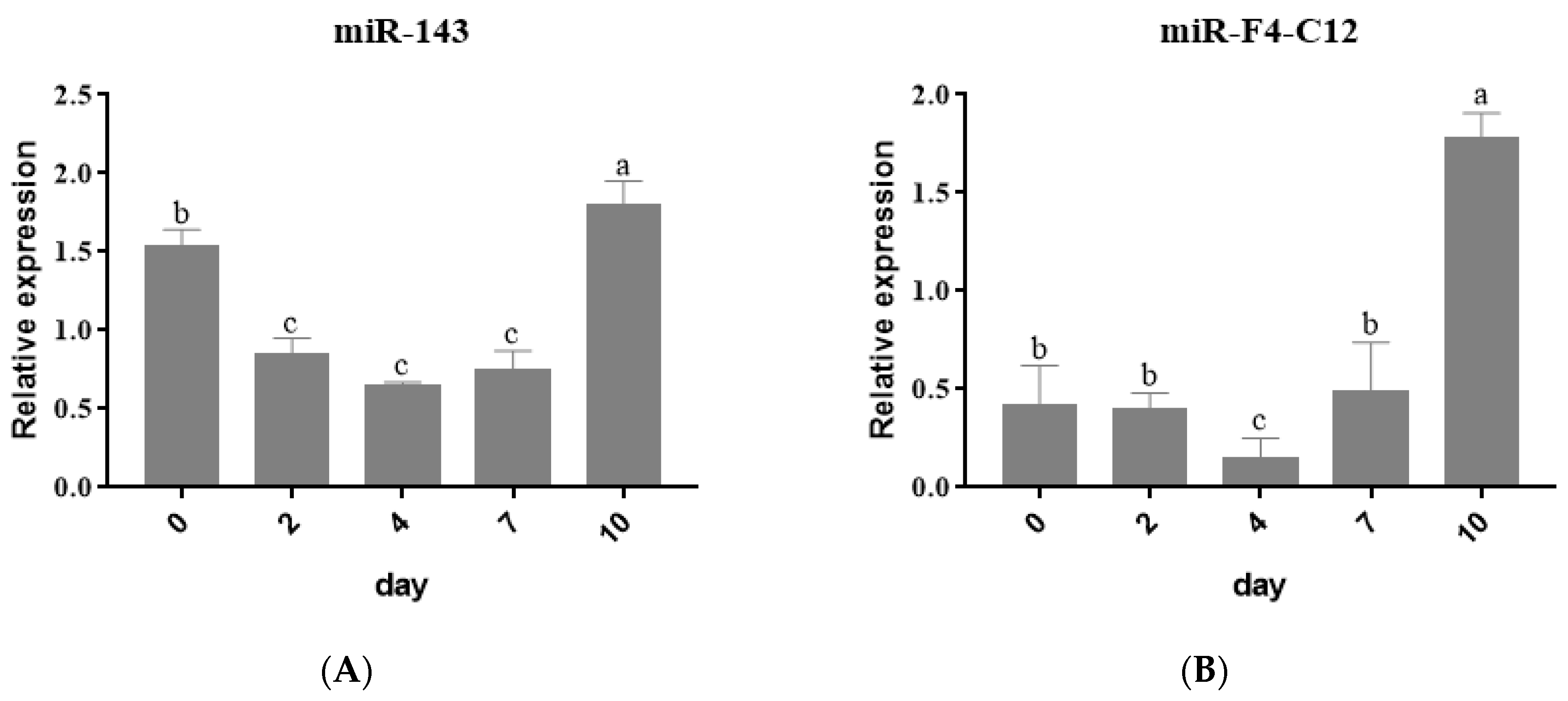
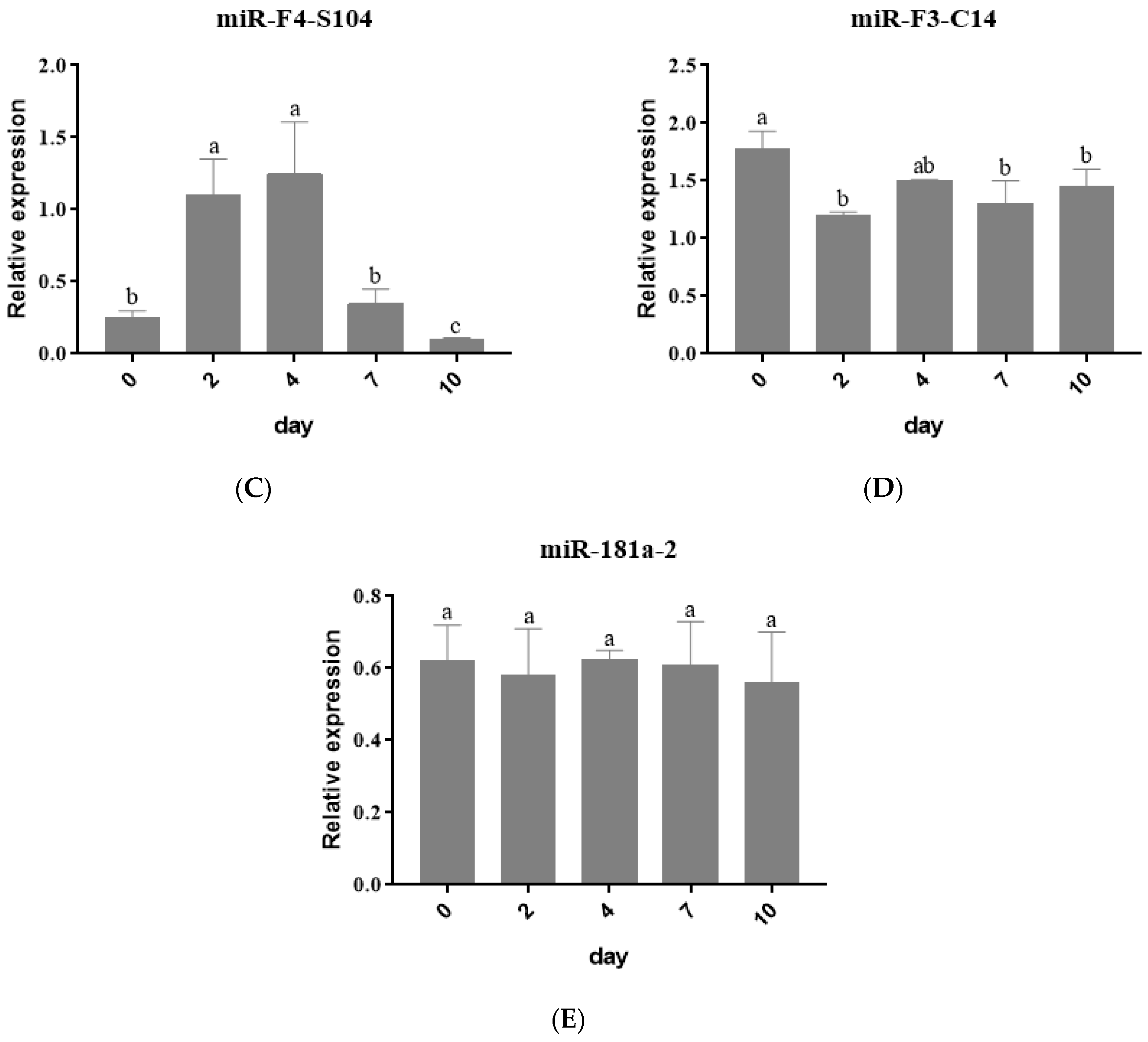
3.2. Expression Profiles of miRNAs during Adipogenic Differentiation in 3T3 Cells
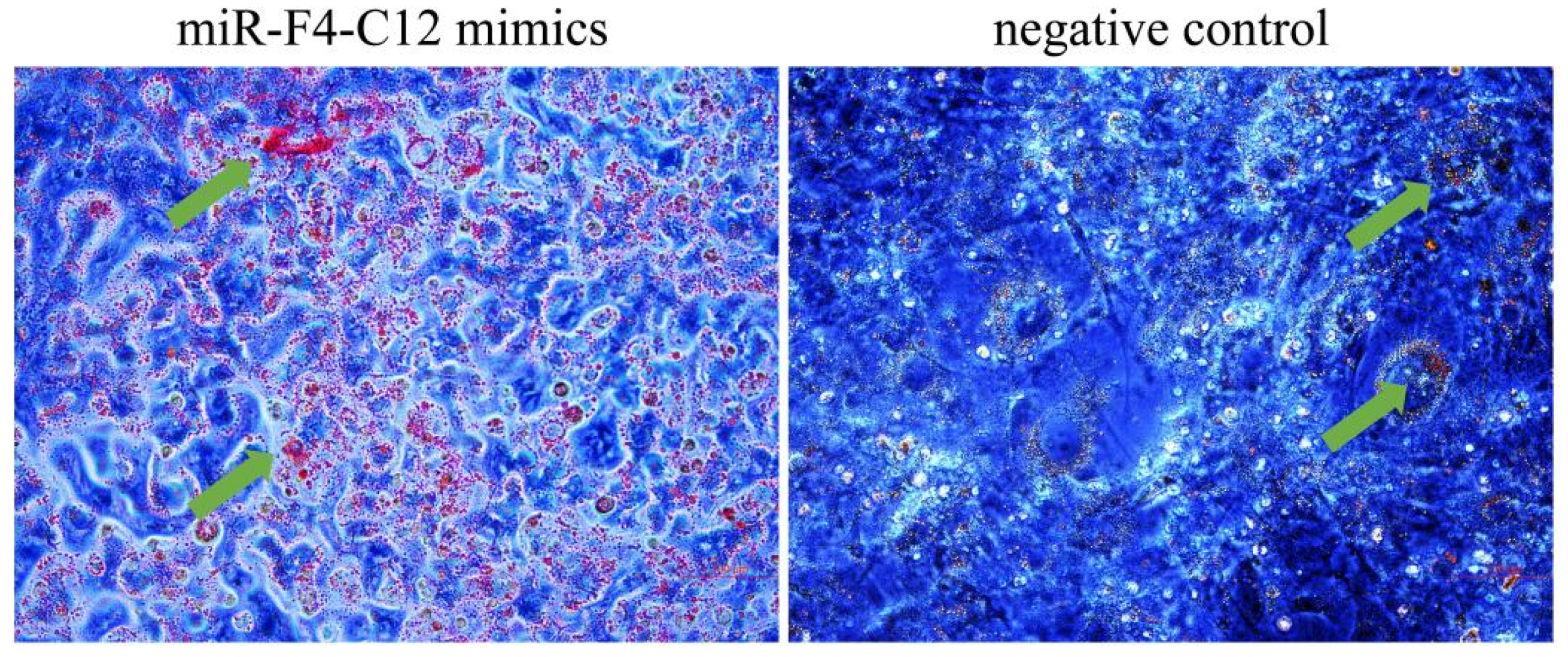
3.3. miR-F4-C12 Promote the Differentiation of 3T3-L1 Cells
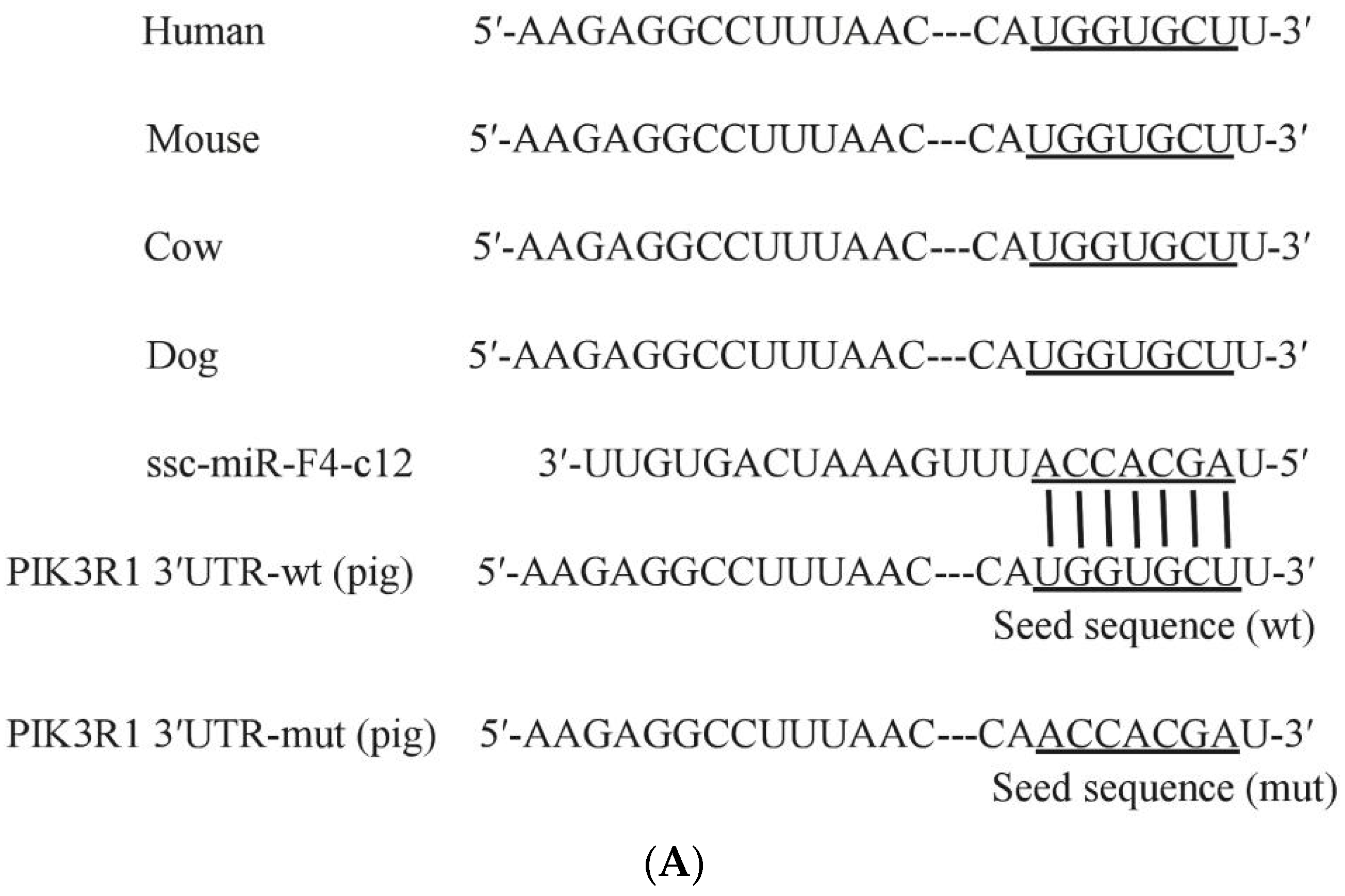
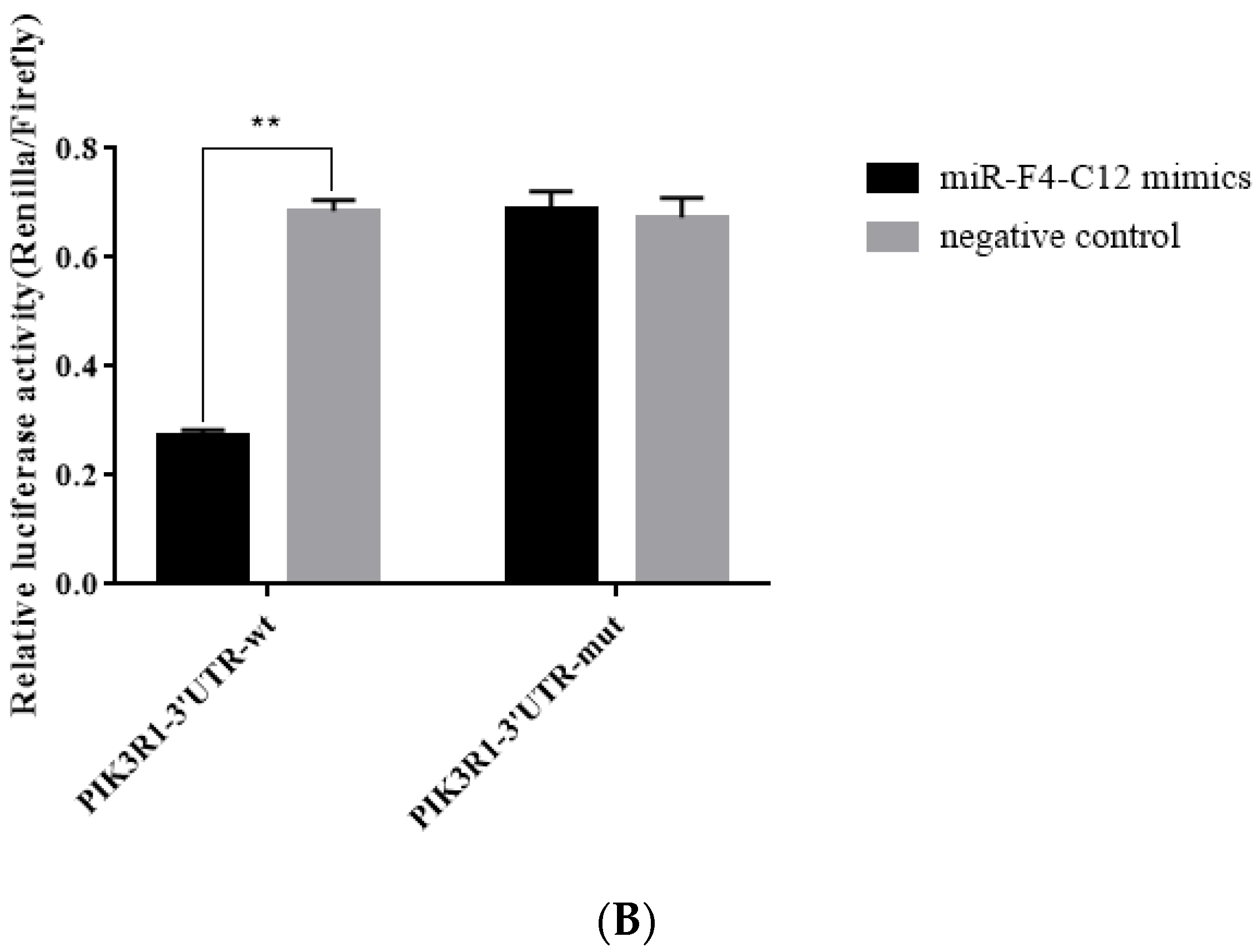
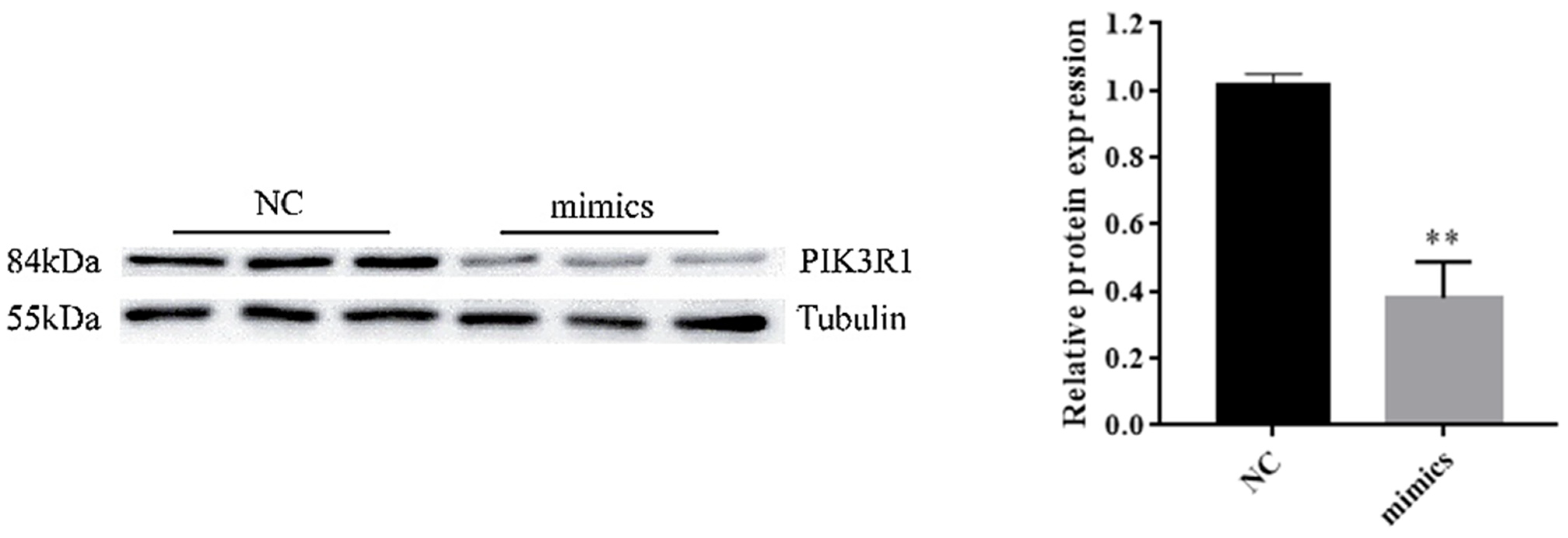
3.4. PIK3R1 Was the Direct Target of miR-F4-C12
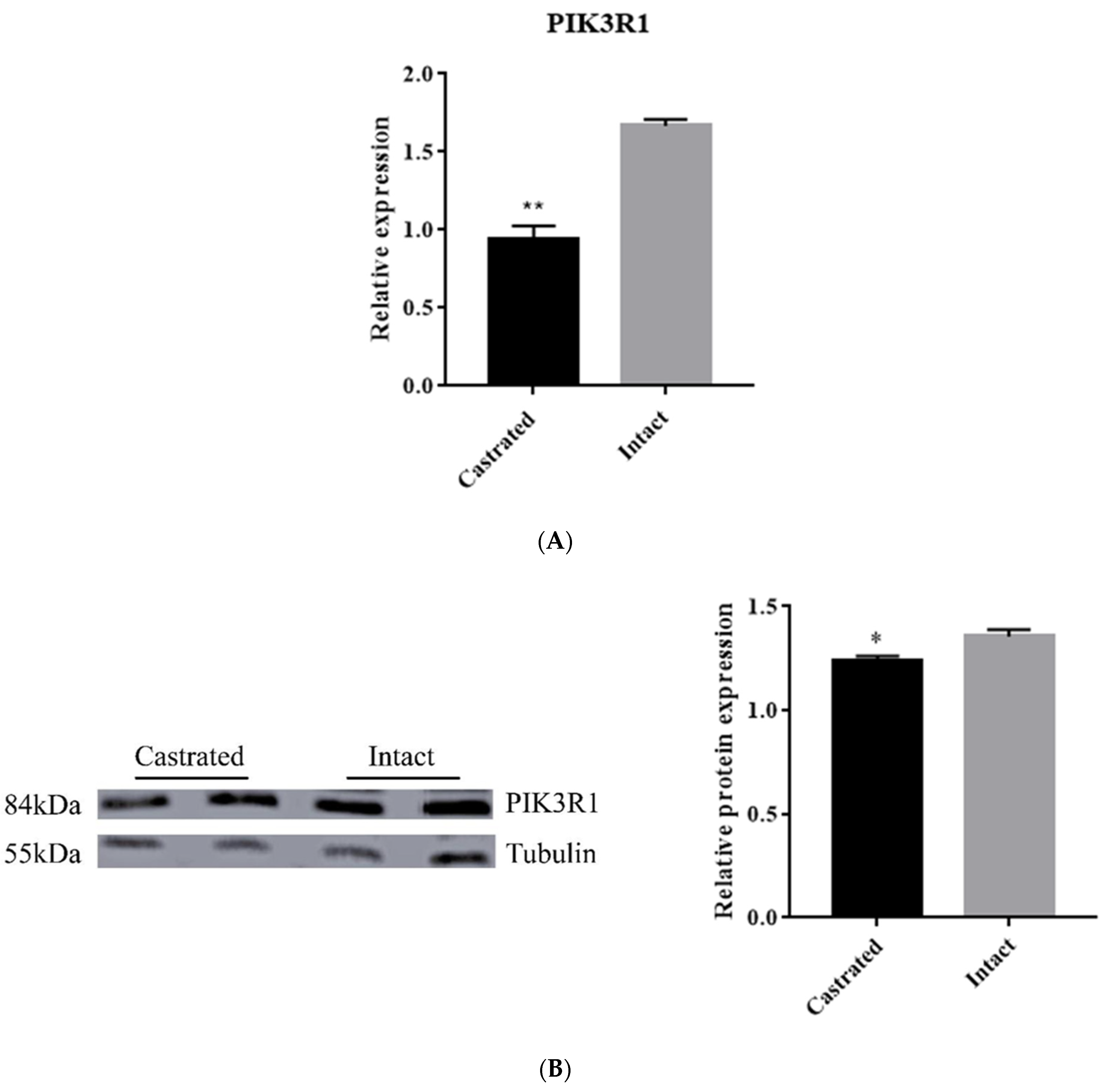
3.5. PIK3R1 Expression in Backfat Tissue
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Christoffersen, B.O.; Gade, L.P.; Golozoubova, V.; Svendsen, O.; Raun, K. Influence of castration-induced testosterone and estradiol deficiency on obesity and glucose metabolism in male Gottingen minipigs. Steroids 2010, 75, 676–684. [Google Scholar] [CrossRef]
- Yao, Y.; Ma, H.; Wu, K.; Shao, Y.; Han, W.; Cai, Z.; Xu, N.; Qi, M.; Zhao, C.; Wu, C. Body composition, serum lipid levels, and transcriptomic characterization in the adipose tissue of male pigs in response to sex hormone deficiency. Gene 2018, 646, 74–82. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Hou, J.; Ye, L.; Chen, Y.; Cui, J.; Tian, W.; Li, C.; Liu, L. MicroRNA-143 regulates adipogenesis by modulating the MAP2K5-ERK5 signaling. Sci. Rep. 2014, 4, 3819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esau, C.; Kang, X.; Peralta, E.; Hanson, E.; Marcusson, E.G.; Ravichandran, L.V.; Sun, Y.; Koo, S.; Perera, R.J.; Jain, R.; et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004, 279, 52361–52365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, P.; Gburcik, V.; Petrovic, N.; Gallagher, I.J.; Nedergaard, J.; Cannon, B.; Timmons, J.A. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr. Disord. 2011, 11, 7. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.J.; Hwang, S.J.; Bae, Y.C.; Jung, J.S. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells 2009, 27, 3093–3102. [Google Scholar] [PubMed]
- Kim, S.Y.; Kim, A.Y.; Lee, H.W.; Son, Y.H.; Lee, G.Y.; Lee, J.W.; Lee, Y.S.; Kim, J.B. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem. Biophys. Res. Commun. 2010, 392, 323–328. [Google Scholar] [CrossRef]
- Karbiener, M.; Fischer, C.; Nowitsch, S.; Opriessnig, P.; Papak, C.; Ailhaud, G.; Dani, C.; Amri, E.Z.; Scheideler, M. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem. Biophys. Res. Commun. 2009, 390, 247–251. [Google Scholar] [CrossRef]
- Peng, Y.; Yu, S.; Li, H.; Xiang, H.; Peng, J.; Jiang, S. MicroRNAs: Emerging roles in adipogenesis and obesity. Cell Signal 2014, 26, 1888–1896. [Google Scholar] [CrossRef] [PubMed]
- Hilton, C.; Neville, M.J.; Karpe, F. MicroRNAs in adipose tissue: Their role in adipogenesis and obesity. Int. J. Obes. 2013, 37, 325–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGregor, R.A.; Choi, M.S. microRNAs in the regulation of adipogenesis and obesity. Curr. Mol. Med. 2011, 11, 304–316. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, Y.; Xu, M.Q.; Wang, C.J.; Fu, X.H.; Liu, J.B.; Han, D.X.; Jiang, H.; Yuan, B.; Zhang, J.B. miR-181a regulate porcine preadipocyte differentiation by targeting TGFBR1. Gene 2019, 681, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Chen, F.F.; Ge, J.; Zhu, J.Y.; Shi, X.E.; Li, X.; Yu, T.Y.; Chu, G.Y.; Yang, G.S. miR-429 Inhibits Differentiation and Promotes Proliferation in Porcine Preadipocytes. Int. J. Mol. Sci. 2016, 17, 2047. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Sun, G.; Yuan, B.; Zhang, L.; Gao, Y.; Jiang, H.; Dai, L.; Zhang, J. miR-375 negatively regulates porcine preadipocyte differentiation by targeting BMPR2. FEBS Lett. 2016, 590, 1417–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, D.; Xi, Q.Y.; Cheng, X.; Dong, T.; Zhu, X.T.; Shu, G.; Wang, L.N.; Jiang, Q.Y.; Zhang, Y.L. miR-146a-5p inhibits TNF-alpha-induced adipogenesis via targeting insulin receptor in primary porcine adipocytes. J. Lipid Res. 2016, 57, 1360–1372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, P.; Mai, Y.; Zhang, Z.; Mi, L.; Wu, G.; Chu, G.; Yang, G.; Sun, S. MiR-15a/b promote adipogenesis in porcine pre-adipocyte via repressing FoxO1. Acta Biochim. Biophys. Sin. 2014, 46, 565–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, S.; Yang, X.; Jia, Y.; Li, R.; Zhao, R. Microvesicle-shuttled miR-130b reduces fat deposition in recipient primary cultured porcine adipocytes by inhibiting PPAR-g expression. J. Cell. Physiol. 2014, 229, 631–639. [Google Scholar] [CrossRef]
- Chen, C.; Deng, B.; Qiao, M.; Zheng, R.; Chai, J.; Ding, Y.; Peng, J.; Jiang, S. Solexa sequencing identification of conserved and novel microRNAs in backfat of Large White and Chinese Meishan pigs. PLoS ONE 2012, 7, e31426. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Li, Y.; Li, X.; Ning, X.; Li, M.; Yang, G. MicroRNA identity and abundance in developing swine adipose tissue as determined by Solexa sequencing. J. Cell. Biochem. 2011, 112, 1318–1328. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Huang, J.M.; Liu, G.; Zhang, J.B.; Wang, J.Y.; Liu, C.K.; Fang, M.Y. A comprehensive microRNA expression profile of the backfat tissue from castrated and intact full-sib pair male pigs. BMC Genom. 2014, 15, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Z.; Zhang, L.; Chen, M.; Jiang, X.; Xu, N. Castration-induced changes in microRNA expression profiles in subcutaneous adipose tissue of male pigs. J. Appl. Genet. 2014, 55, 259–266. [Google Scholar] [CrossRef]
- Zhang, X.M.; Wang, L.H.; Su, D.J.; Zhu, D.; Li, Q.M.; Chi, M.H. MicroRNA-29b promotes the adipogenic differentiation of human adipose tissue-derived stromal cells. Obesity 2016, 24, 1097–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Li, M.; Guan, J.; Li, P.; Wang, H.; Guo, Y.; Shuai, S.; Li, X. MicroRNAs miR-27a and miR-143 regulate porcine adipocyte lipid metabolism. Int. J. Mol. Sci. 2011, 12, 7950–7959. [Google Scholar] [CrossRef]
- Zhang, X.; Price, N.L.; Fernandez-Hernando, C. Non-coding RNAs in lipid metabolism. Vascul. Pharmacol. 2019, 114, 93–102. [Google Scholar] [CrossRef]
- Desgagne, V.; Bouchard, L.; Guerin, R. microRNAs in lipoprotein and lipid metabolism: From biological function to clinical application. Clin. Chem. Lab. Med. 2017, 55, 667–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, A.; Zhu, L.; Gupta, N.; Chang, Y.; Fang, F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol. Endocrinol. 2007, 21, 2785–2794. [Google Scholar] [CrossRef] [PubMed]
- Ru, P.; Guo, D. microRNA-29 mediates a novel negative feedback loop to regulate SCAP/SREBP-1 and lipid metabolism. RNA Dis. 2017, 4, e1525. [Google Scholar]
- Yu, W.; Chen, Z.; Zhang, J.; Zhang, L.; Ke, H.; Huang, L.; Peng, Y.; Zhang, X.; Li, S.; Lahn, B.T.; et al. Critical role of phosphoinositide 3-kinase cascade in adipogenesis of human mesenchymal stem cells. Mol. Cell. Biochem. 2008, 310, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Terauchi, Y.; Matsui, J.; Kamon, J.; Yamauchi, T.; Kubota, N.; Komeda, K.; Aizawa, S.; Akanuma, Y.; Tomita, M.; Kadowaki, T. Increased serum leptin protects from adiposity despite the increased glucose uptake in white adipose tissue in mice lacking p85alpha phosphoinositide 3-kinase. Diabetes 2004, 53, 2261–2270. [Google Scholar] [CrossRef] [Green Version]
- Kuo, T.; Chen, T.C.; Lee, R.A.; Nguyen, N.; Broughton, A.E.; Zhang, D.; Wang, J.C. Pik3r1 Is Required for Glucocorticoid-Induced Perilipin 1 Phosphorylation in Lipid Droplet for Adipocyte Lipolysis. Diabetes 2017, 66, 1601–1610. [Google Scholar] [CrossRef] [Green Version]
| Primer Name | Primer Sequence (5′-3′) | Application |
|---|---|---|
| miR-F4-C12 | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACACTGA | qRT-PCR |
| F: TCTCGCACGC TAGCACCATT | ||
| R: CTCAACTGGTGTCGTGGAGTC | ||
| miR-F3-C14 | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGCCAATA | qRT-PCR |
| F: GGG TAGCAGCACGTAAATATT | ||
| R: CTCAACTGGTGTCGTGGAGTC | ||
| miR-F4-S104 | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTTCAGTCA | qRT-PCR |
| F: GGG CATCTGTGGGATTATGA | ||
| R: CTCAACTGGTGTCGTGGAGTC | ||
| miR-181a-2 | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACTCACC | qRT-PCR |
| F: TCTCGCACGC AACATTCAACGCTG | ||
| R: CTCAACTGGTGTCGTGGAGTC | ||
| miR-143 | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGAGCTACA | qRT-PCR |
| F: TCTCGCACGCTGAGATGAAGC | ||
| R: CTCAACTGGTGTCGTGGAGTC | ||
| PIK3R1 | F: GACTTGCCCCACCACGAT | qRT-PCR |
| R: AGTGCTTGACCTCGCCGT | ||
| PIK3R1-wt-3′ UTR | F: ATTGCGATCGCGCCACCTGCTCAAGTTCA | Luciferase reporter |
| R: ATTGTTTAAACGTTCCCCAAAGTGTTCCTCT | ||
| PIK3R1-mut- 3′ UTR | F: ATTGCGATCGCGCCACCTGCTCAAGTTCA | Luciferase reporter |
| R: ATTGTTTAAACGCTTCTGAAAGCATGAACATCGTGGTTGGTGAAAGGCCTCTTTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Q.; Chen, J.; Liu, X.; Luo, Y.; Wang, T.; Fang, M. miR-F4-C12 Functions on the Regulation of Adipose Accumulation by Targeting PIK3R1 in Castrated Male Pigs. Animals 2021, 11, 3053. https://doi.org/10.3390/ani11113053
Xu Q, Chen J, Liu X, Luo Y, Wang T, Fang M. miR-F4-C12 Functions on the Regulation of Adipose Accumulation by Targeting PIK3R1 in Castrated Male Pigs. Animals. 2021; 11(11):3053. https://doi.org/10.3390/ani11113053
Chicago/Turabian StyleXu, Qiao, Jie Chen, Ximing Liu, Yabiao Luo, Tianzuo Wang, and Meiying Fang. 2021. "miR-F4-C12 Functions on the Regulation of Adipose Accumulation by Targeting PIK3R1 in Castrated Male Pigs" Animals 11, no. 11: 3053. https://doi.org/10.3390/ani11113053
APA StyleXu, Q., Chen, J., Liu, X., Luo, Y., Wang, T., & Fang, M. (2021). miR-F4-C12 Functions on the Regulation of Adipose Accumulation by Targeting PIK3R1 in Castrated Male Pigs. Animals, 11(11), 3053. https://doi.org/10.3390/ani11113053





