Propionibacterium acnes and Acne Vulgaris: New Insights from the Integration of Population Genetic, Multi-Omic, Biochemical and Host-Microbe Studies
Abstract
:1. Introduction
2. Taxonomy and Intraspecies Diversity of Propionibacterium acnes
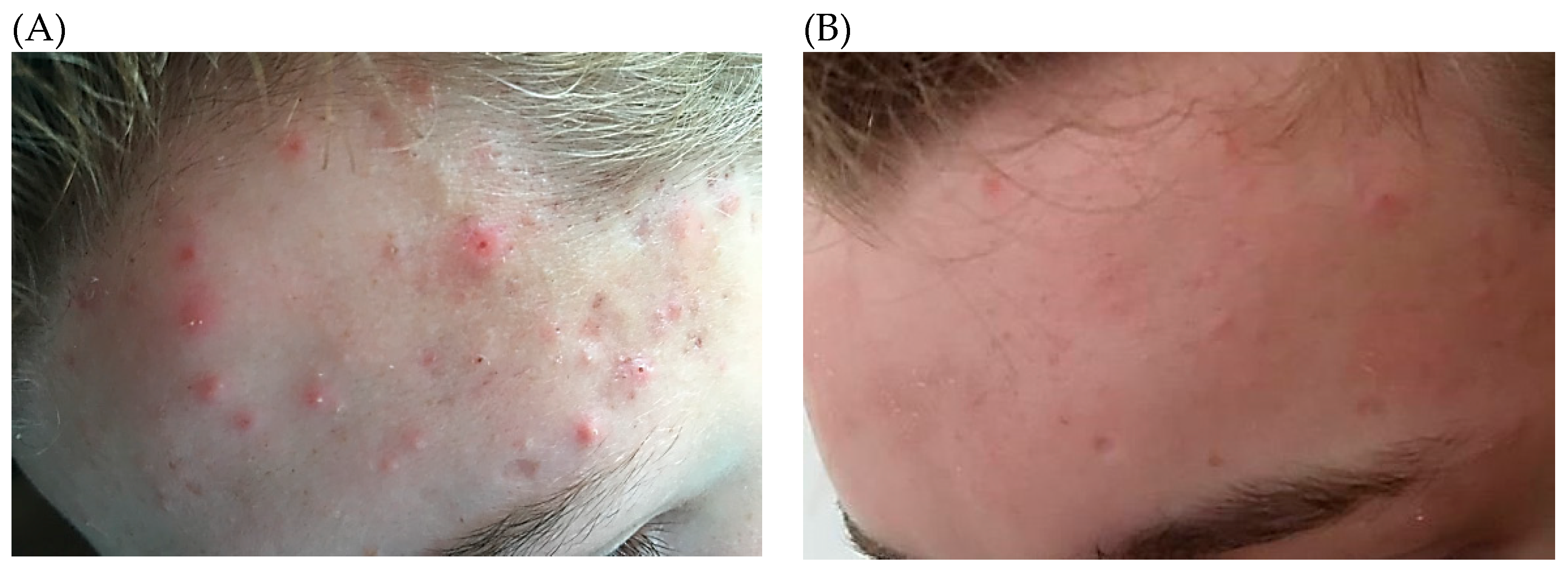
3. Acne Vulgaris
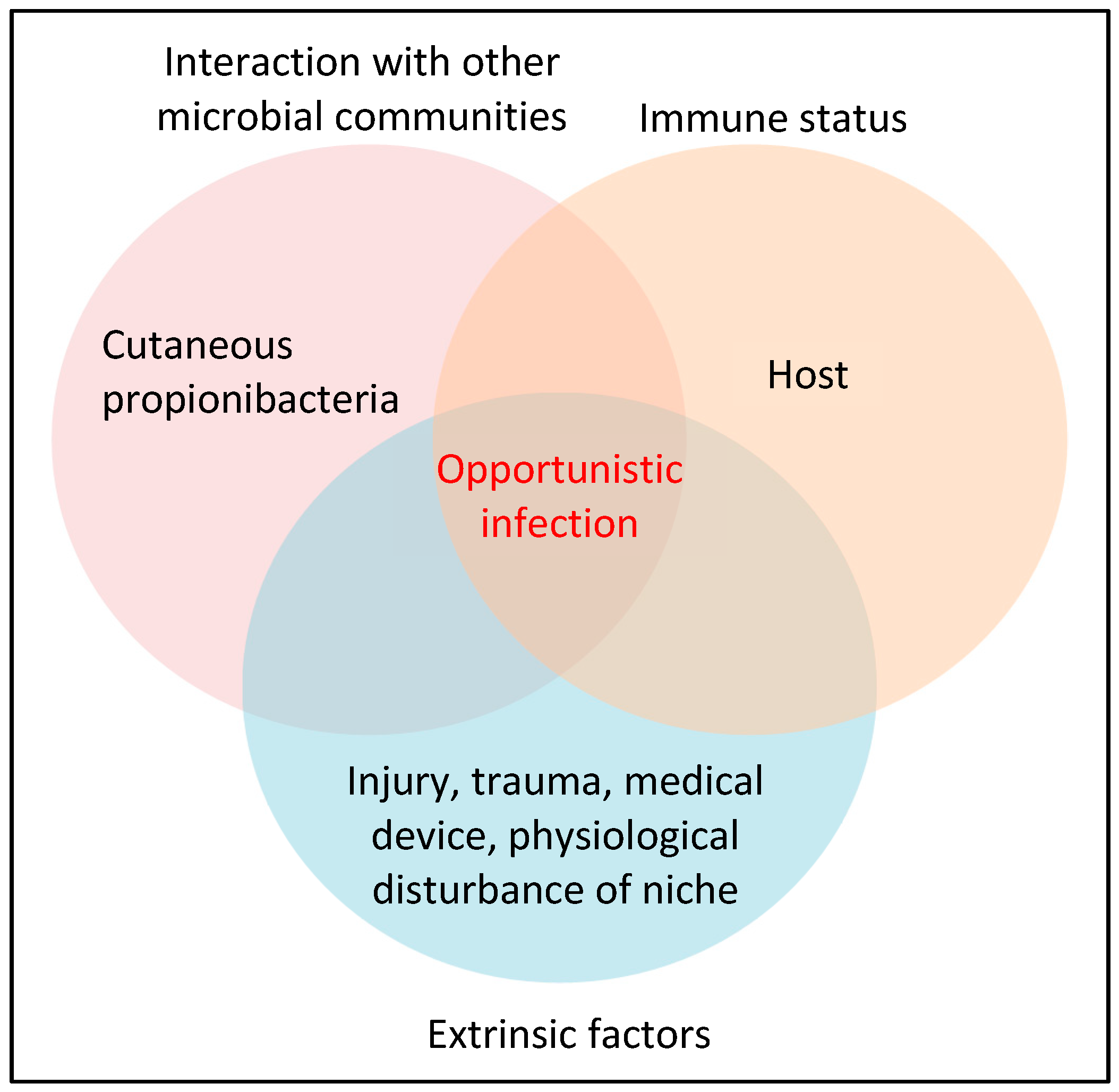
3.1. What is the Evidence that P. acnes is a Pathogenic Factor in Acne Development?
3.2. Inflammatory Responses in Acne: How does P. acnes Fit the Bill?
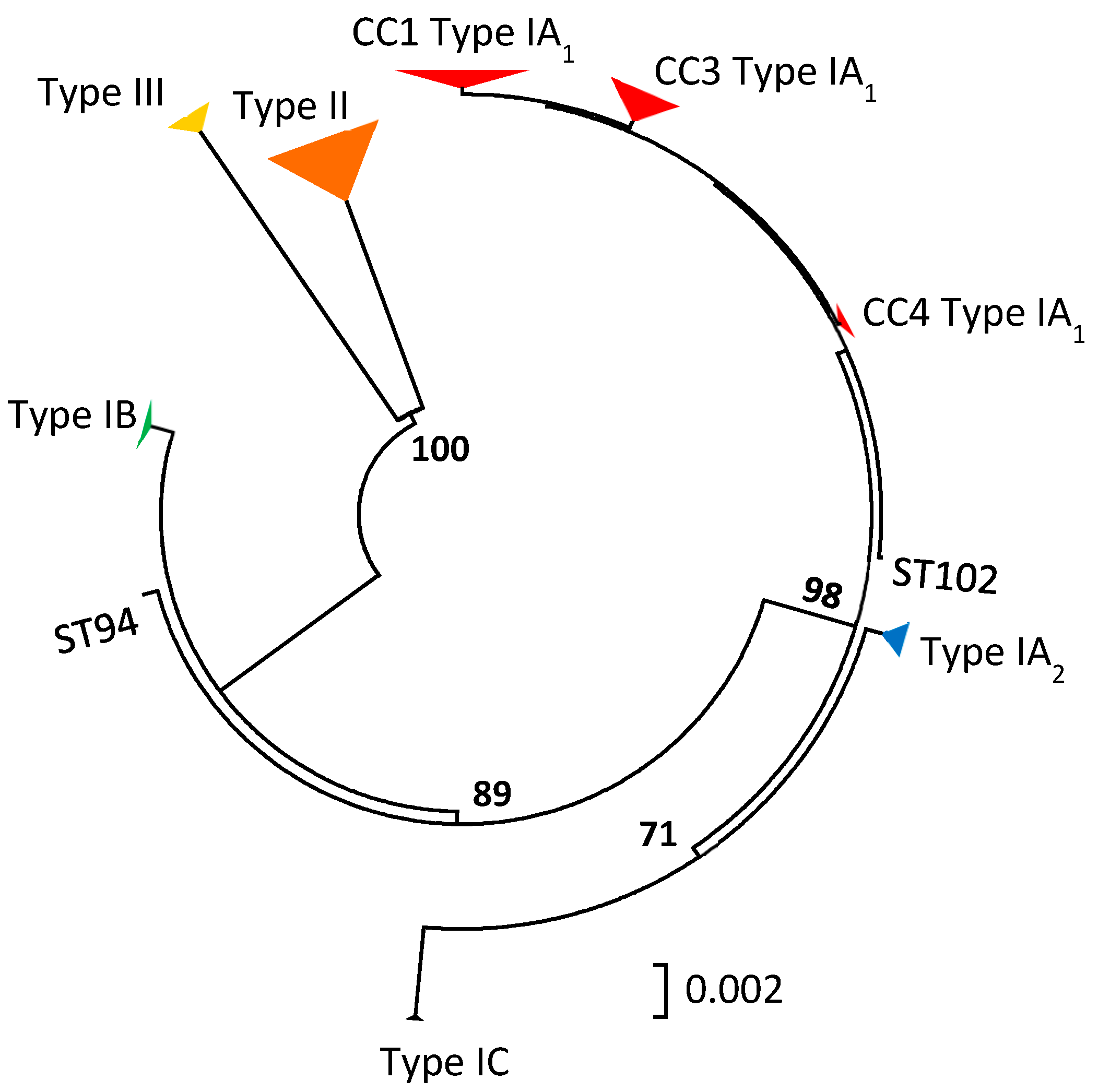
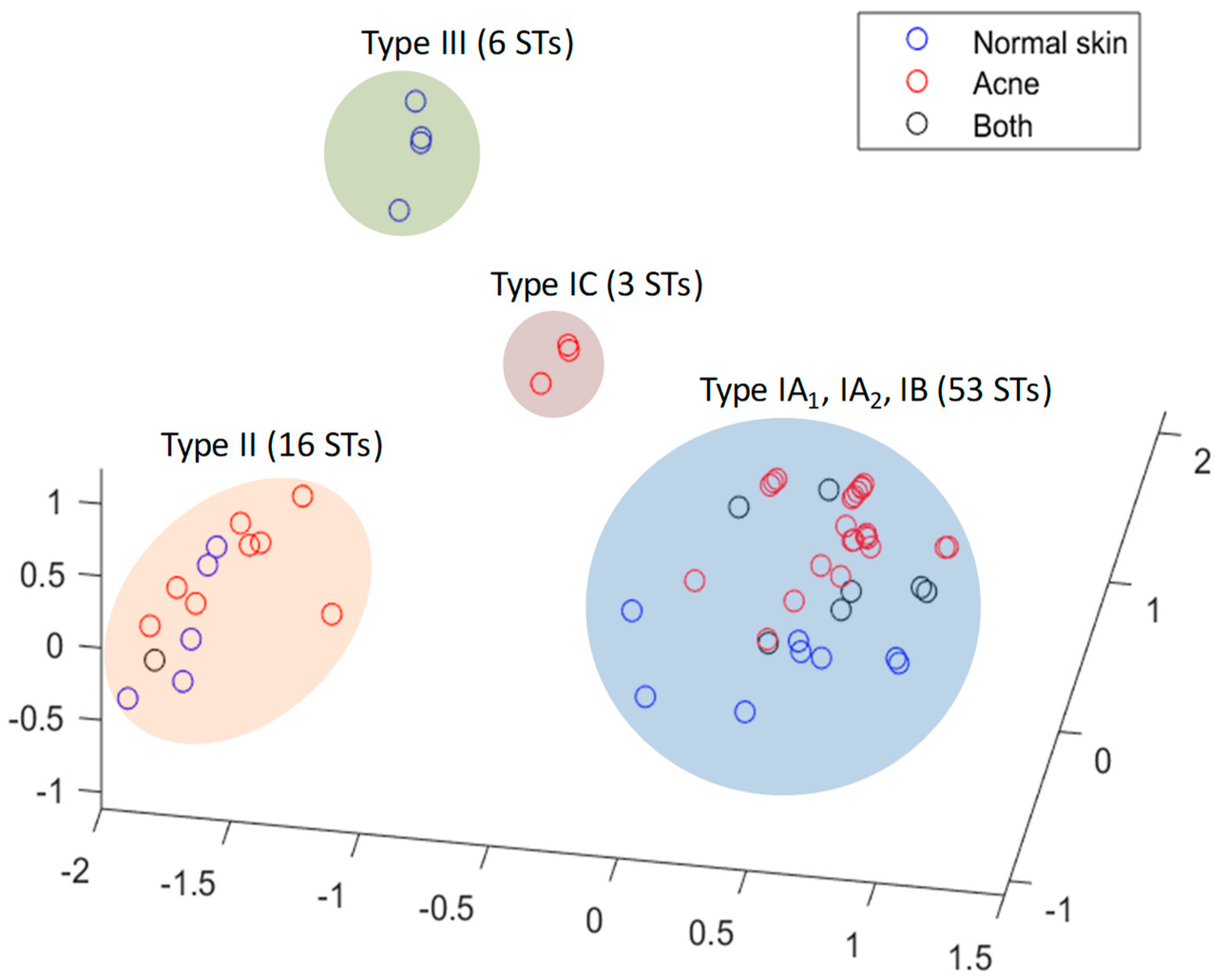
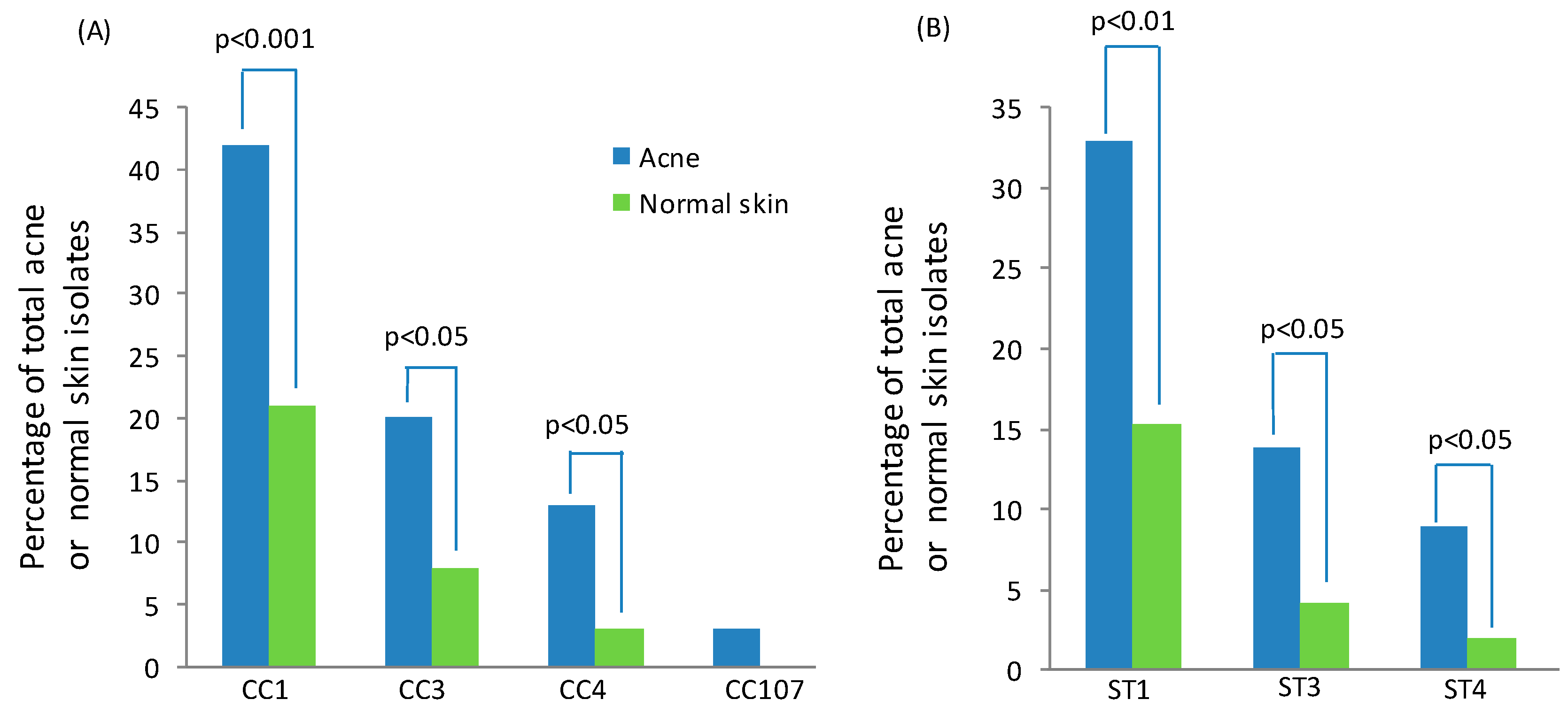
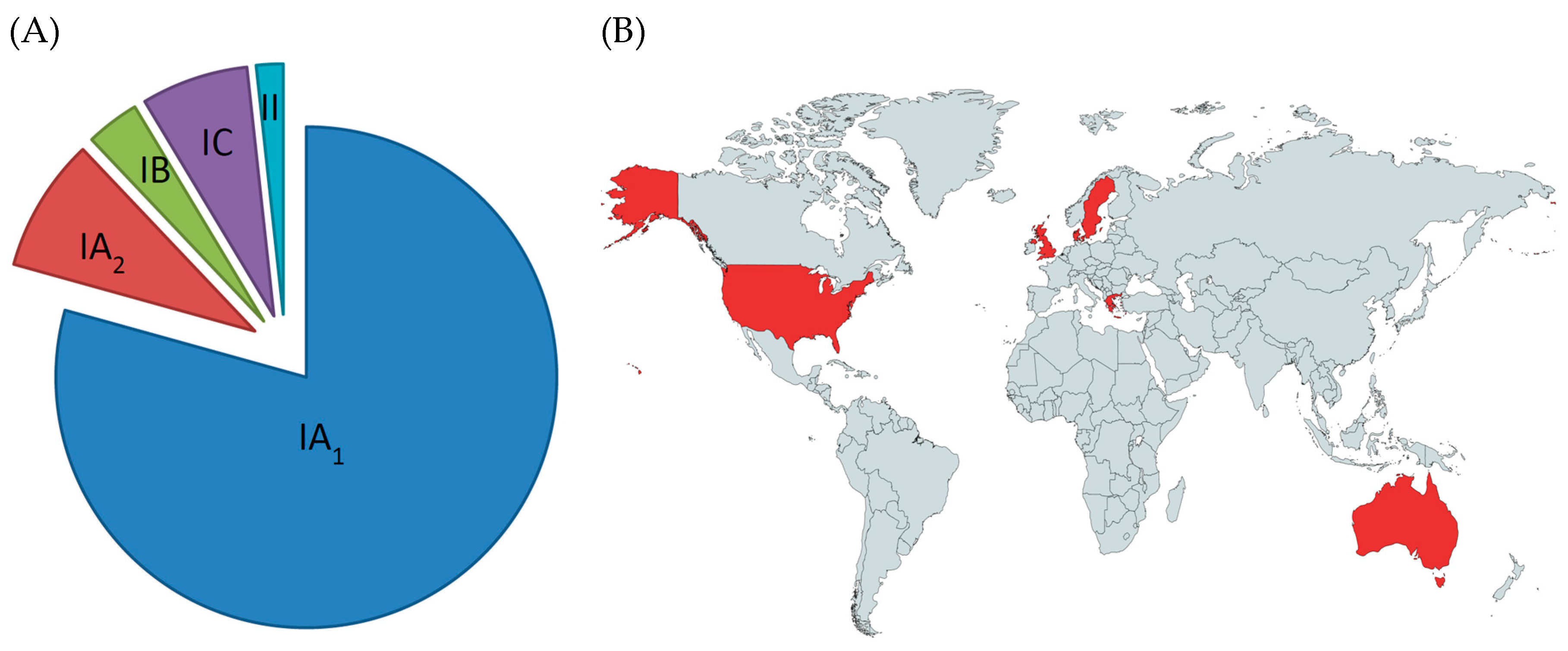
3.3. Insights into the Role of P. acnes in Acne from Population Genetic Analyses
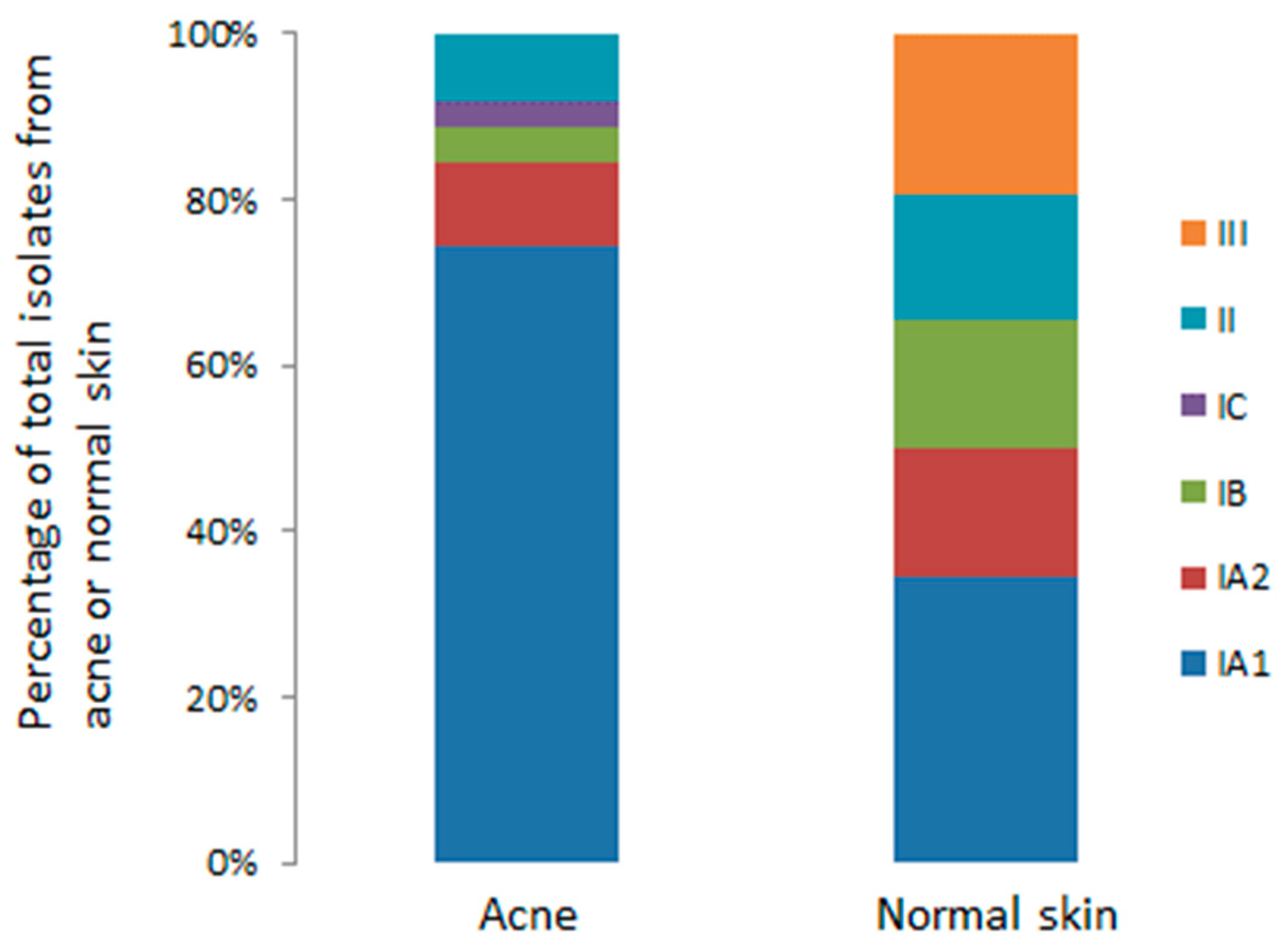
3.3.1. Culture-Based Analysis
3.3.2. 16S rDNA Metagenomic Analysis
3.3.3. Shotgun Metagenomic Analysis
3.3.4. Antibiotic Resistance
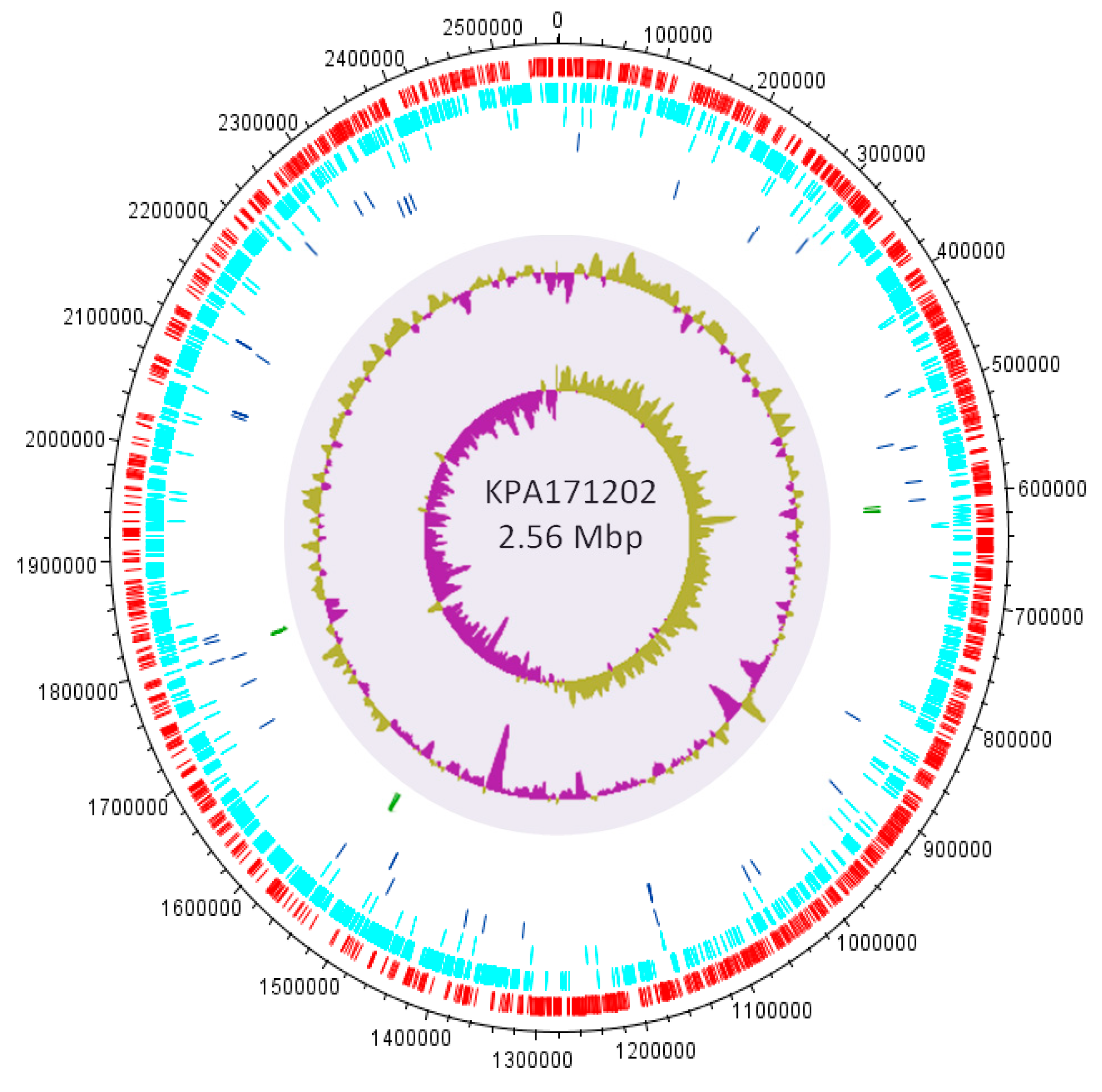
3.4. Insights into the Role of P. acnes in Acne from Comparative Genomics
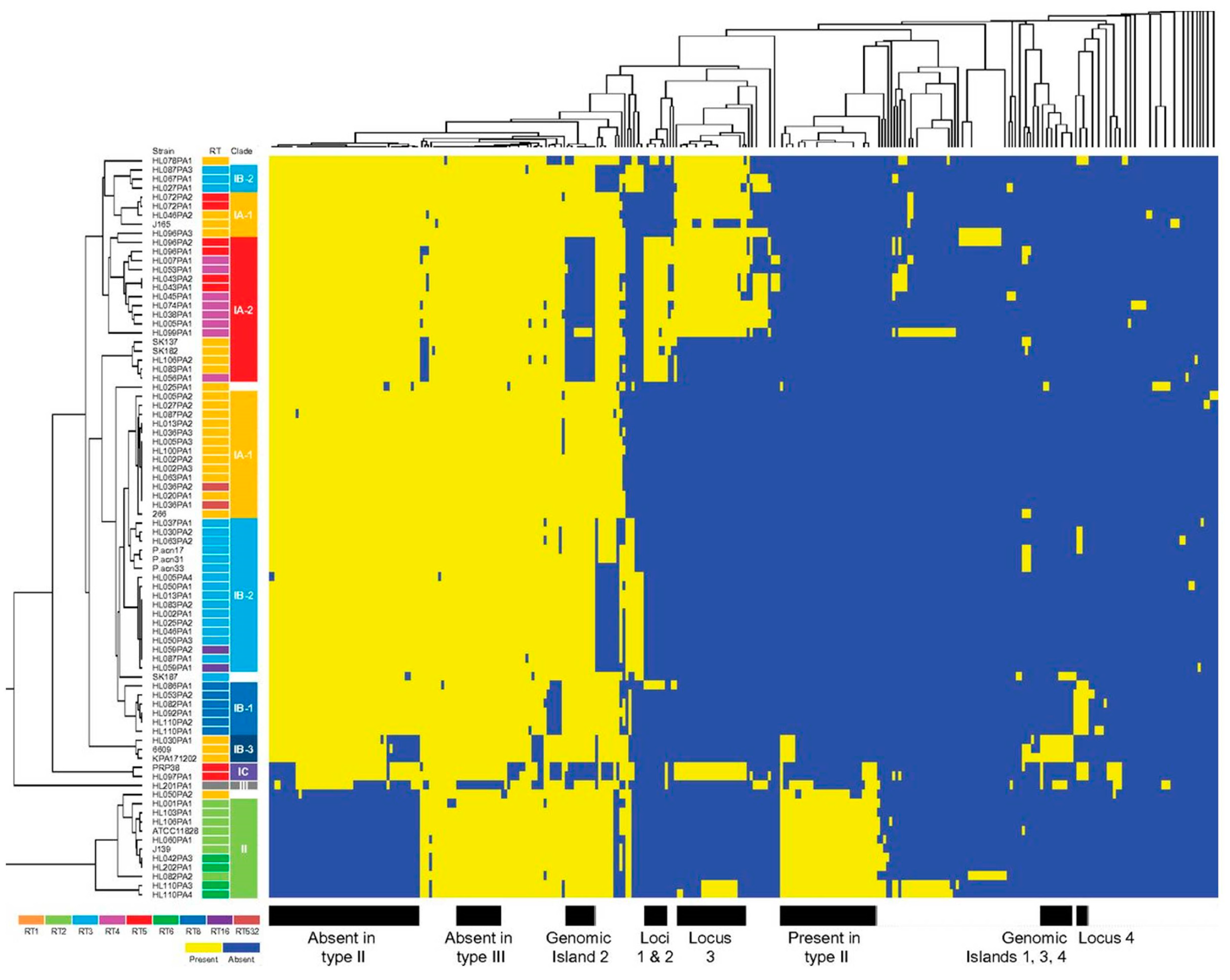
3.4.1. Genomic Islands and a Flexible Gene Pool
3.4.2. CRISPR/Cas
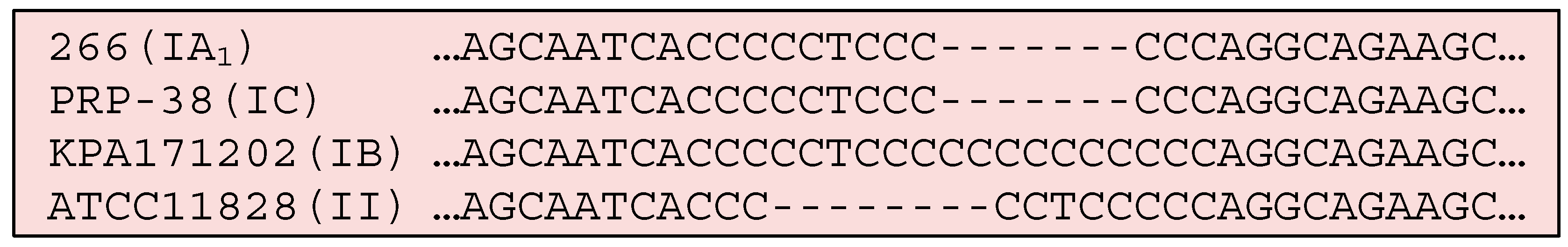
3.4.3. Homopolymeric Tracts (HPTs)
3.5. Insights into the Role of P. acnes in Acne from Transcriptomic and Proteomic Analysis of Phylogroups
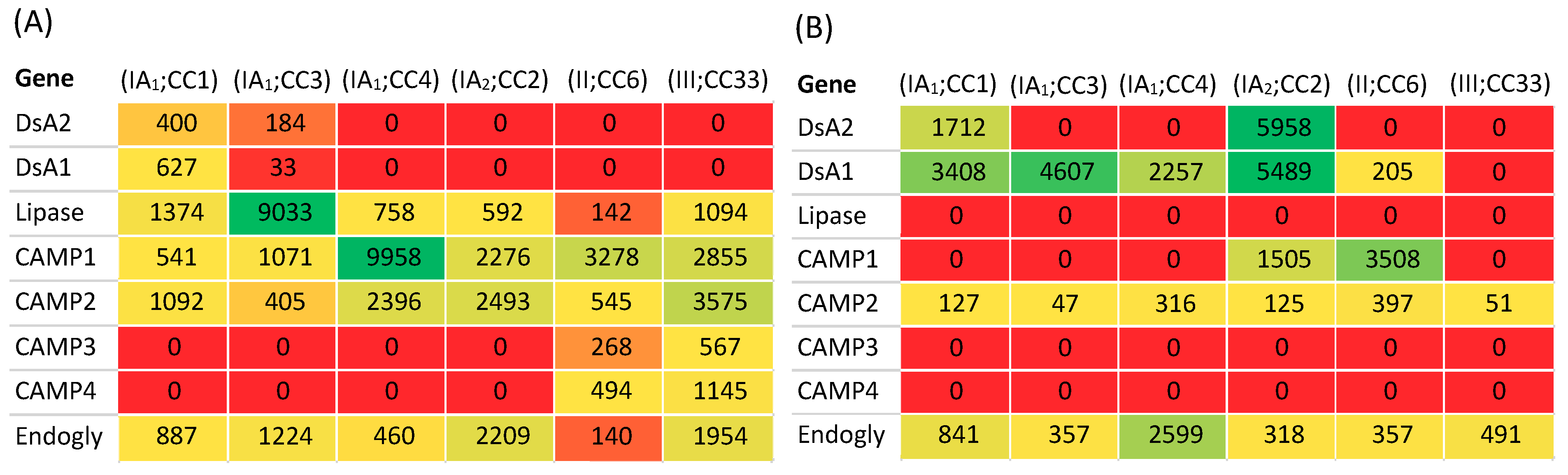
3.5.1. Transcriptome
3.5.2. Proteome
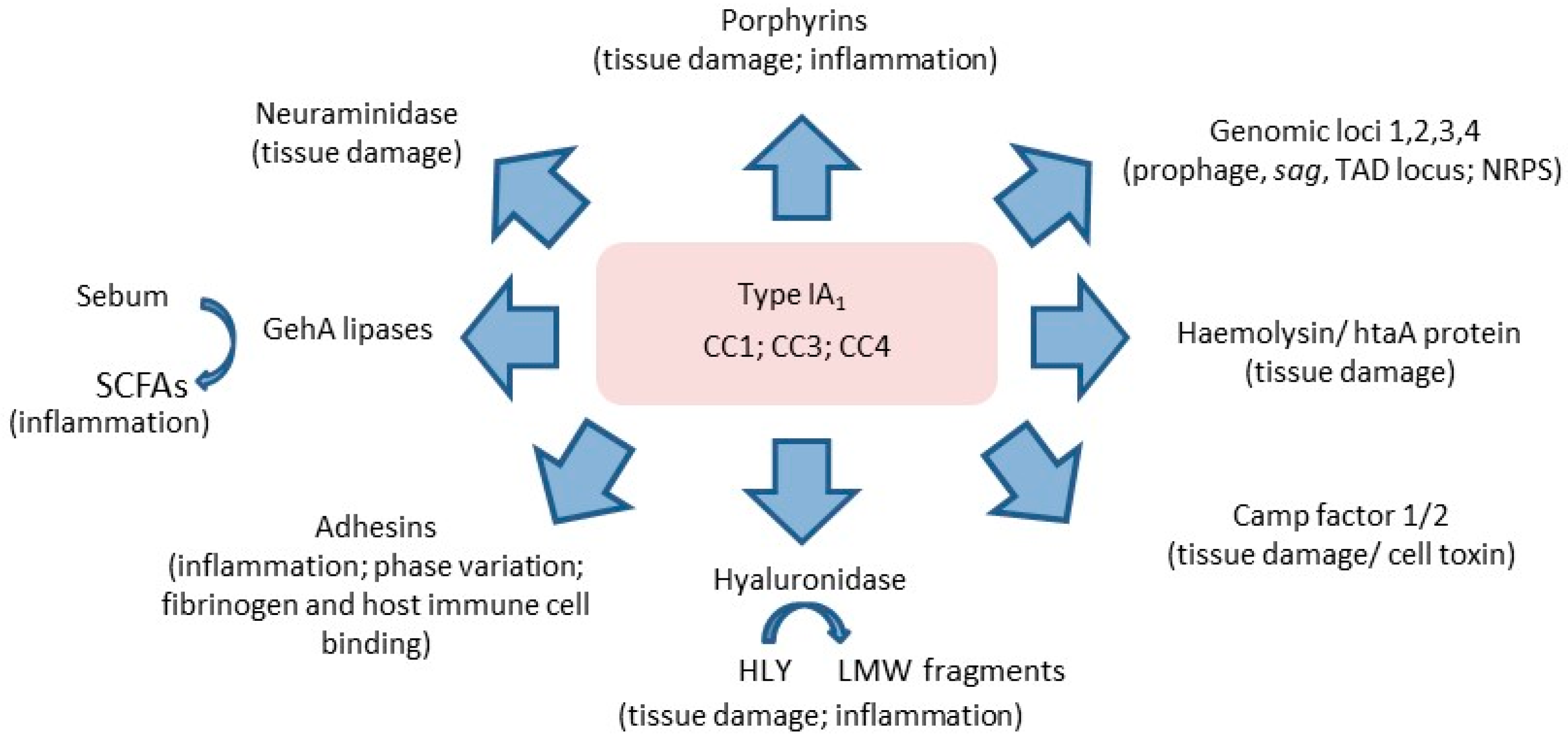
3.6. Insights into the Role of P. acnes in Acne from Other Biochemical Studies of Phylogroups
3.7. Insights into the Role of P. acnes in Acne from Cell-Based and Skin-Explant Studies
3.8. Insights into the Role of P. acnes in Acne from Meta-Transcriptomic and Meta-Proteomic Analysis of Follicular Microbiota
3.9. New Therapeutic Strategies
3.9.1. Bacteriophage Therapy
3.9.2. Skin Probiotics
3.9.3. Vaccine Development
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barnard, E.; Li, H. Shaping of cutaneous function by encounters with commensals. J. Physiol. 2017, 595, 437–450. [Google Scholar] [CrossRef]
- Shu, M.; Wang, Y.; Yu, J.; Kuo, S.; Coda, A.; Jiang, Y.; Gallo, R.L.; Huang, C.-M. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS ONE 2013, 8, e55380. [Google Scholar] [CrossRef] [PubMed]
- Christensen, G.J.M.; Scholz, C.F.P.; Enghild, J.; Rohde, H.; Kilian, M.; Thürmer, A.; Brzuszkiewicz, E.; Lomholt, H.B.; Brüggemann, H. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genom. 2016, 15, 152. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, S.; Nakamura, T. Purification and properties of a bacteriocin-like substance (acnecin) of oral Propionibacterium acnes. Antimicrob. Agents Chemother. 1978, 14, 893–898. [Google Scholar] [CrossRef] [PubMed]
- Perry, A.; Lambert, P. Propionibacterium acnes: Infection beyond the skin. Expert Rev. Anti. Infect. Ther. 2011, 12, 1149–1156. [Google Scholar] [CrossRef]
- McGinley, K.J.; Webster, G.F.; Leyden, J.J. Regional variations of cutaneous propionibacteria. Appl. Environ. Microbiol. 1979, 35, 62–66. [Google Scholar]
- Chang, H.W.; Yan, D.; Singh, R.; Liu, J.; Lu, X.; Ucmak, D.; Lee, K.; Afifi, L.; Fadrosh, D.; Leech, J.; et al. Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome 2018, 6, 154. [Google Scholar] [CrossRef]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; NISC Comparative Sequence Program; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Beylot, C.; Auffret, N.; Poli, F.; Claudel, J.-P.; Leccia, M.-T.; Del Giudice, P.; Dreno, B. Propionibacterium acnes: An update on its role in the pathogenesis of acne. J. Eur. Acad. Dermatol. Venereol. 2014, 28, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Barnard, E.; Liu, J.; Yankova, E.; Cavalcanti, S.M.; Magalhães, M.; Li, H.; Patrick, S.; McDowell, A. Strains of the Propionibacterium acnes type III lineage are associated with the skin condition progressive macular hypomelanosis. Sci. Rep. 2016, 6, 3196. [Google Scholar] [CrossRef]
- Petersen, R.L.; Scholz, C.F.; Jensen, A.; Brüggemann, H.; Lomholt, H.B. Propionibacterium acnes phylogenetic type III is associated with progressive macular hypomelanosis. Eur. J. Microbiol. Immunol. 2017, 7, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Piper, K.E.; Jacobson, M.J.; Cofield, R.H.; Sperling, J.W.; Sanchez-Sotelo, J.; Osmon, D.R.; McDowell, A.; Patrick, S.; Steckelberg, J.M.; Mandrekar, J.N.; et al. Microbiologic diagnosis of prosthetic shoulder infection by use of implant sonication. J. Clin. Microbiol. 2009, 47, 1878–1884. [Google Scholar] [CrossRef] [PubMed]
- Niazi, S.A.; Clarke, D.; Do, T.; Gilbert, S.C.; Mannocci, F.; Beighton, D. Propionibacterium acnes and Staphylococcus epidermidis isolated from refractory endodontic lesions are opportunistic pathogens. J. Clin. Microbiol. 2010, 48, 3859–3869. [Google Scholar] [CrossRef]
- Eishi, Y. Etiologic link between sarcoidosis and Propionibacterium acnes. Respir. Investig. 2013, 51, 56–68. [Google Scholar] [CrossRef]
- Albert, H.B.; Lambert, P.; Rollason, J.; Sorensen, J.S.; Worthington, T.; Pedersen, M.B.; Nørgaard, H.S.; Vernallis, A.; Busch, F.; Manniche, C.; et al. Does nuclear tissue infected with bacteria following disc herniations lead to Modic changes in the adjacent vertebrae? Eur. Spine J. 2013, 22, 690–696. [Google Scholar] [CrossRef]
- Cohen, R.J.; Shannon, B.A.; McNeal, J.E.; Shannon, T.; Garrett, K.L. Propionibacterium acnes associated with inflammation in radical prostatectomy specimens: A possible link to cancer evolution? J. Urol. 2005, 173, 1969–1974. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.W.; Li, C.Y.; Zhao, X.D.; Liu, Y.F. Fatal bacteria granuloma after trauma: A new entity. Br. J. Dermatol. 2002, 147, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Zaffiri, L.; Abdulmassih, R.; Boyaji, S.; Bagh, I.; Campbell, A.R.; Loehrke, M.E. Brain abscess induced by Propionibacterium acnes in a patient with severe chronic sinusitis. New Microbiol. 2013, 36, 325–329. [Google Scholar]
- McDowell, A.; Valanne, S.; Ramage, G.; Tunney, M.M.; Glenn, J.V.; McLorinan, G.C.; Bhatia, A.; Maisonneuve, J.F.; Lodes, M.; Persing, D.H.; et al. Propionibacterium acnes types I and II represent phylogenetically distinct groups. J. Clin. Microbiol. 2005, 43, 326–334. [Google Scholar] [CrossRef]
- McDowell, A.; Perry, A.L.; Lambert, P.A.; Patrick, S. A new phylogenetic group of Propionibacterium acnes. J. Med. Microbiol. 2008, 57, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Lomholt, H.B.; Kilian, M. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS ONE 2010, 5, e12277. [Google Scholar] [CrossRef]
- McDowell, A.; Barnard, E.; Nagy, I.; Gao, A.; Tomida, S.; Li, H.; Eady, A.; Cove, J.; Nord, C.E.; Patrick, S. An expanded multilocus sequence typing scheme for Propionibacterium acnes: Investigation of “pathogenic”, “commensal” and antibiotic resistant strains. PLoS ONE 2012, 7, e41480. [Google Scholar] [CrossRef]
- Dekio, I.; Culak, R.; Misra, R.; Gaulton, T.; Fang, M.; Sakamoto, M.; Ohkuma, M.; Oshima, K.; Hattori, M.; Klenk, H.P.; et al. Dissecting the taxonomic heterogeneity within Propionibacterium acnes: Proposal for Propionibacterium acnes subsp. acnes subsp. nov. and Propionibacterium acnes subsp. elongatum subsp. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 4776–4787. [Google Scholar] [CrossRef]
- McDowell, A.; Barnard, E.; Liu, J.; Li, H.; Patrick, S. Emendation of Propionibacterium acnes subsp. acnes (Deiko et al. 2015) and proposal of Propionibacterium acnes type II as Propionibacterium acnes subsp. defendens subsp. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5358–5365. [Google Scholar]
- Scholz, C.F.; Kilian, M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 4422–4432. [Google Scholar] [CrossRef]
- McDowell, A. Over a decade of recA and tly gene sequence typing of the skin bacterium Propionibacterium acnes: What have we learnt? Microorganisms 2017, 6, 1. [Google Scholar] [CrossRef]
- McDowell, A.; Nagy, I.; Magyari, M.; Barnard, E.; Patrick, S. The opportunistic pathogen Propionibacterium acnes: Insights into typing, human disease, clonal diversification and CAMP factor evolution. PLoS ONE 2013, 8, e70897. [Google Scholar] [CrossRef]
- Nagy, I.; Pivarcsi, A.; Koreck, A.; Széll, M.; Urbán, E.; Kemény, L. Distinct strains of Propionibacterium acnes induce selective human β-defensin-2 and interleukin-8 expression in human keratinocytes through toll-like receptors. J. Investig. Dermatol. 2005, 124, 931–938. [Google Scholar] [CrossRef]
- Nagy, I.; Pivarcsi, A.; Kis, K.; Koreck, A.; Bodai, L.; McDowell, A.; Seltmann, H.; Patrick, S.; Zouboulis, C.C.; Kemény, L. Propionibacterium acnes and lipopolysaccharide induce the expression of antimicrobial peptides and proinflammatory cytokines/chemokines in human sebocytes. Microbes Infect. 2006, 8, 2195–2205. [Google Scholar] [CrossRef]
- Valanne, S.; McDowell, A.; Ramage, G.; Tunney, M.M.; Einarsson, G.G.; O’Hagan, S.; Wisdom, G.B.; Fairley, D.; Bhatia, A.; Maisonneuve, J.F.; et al. CAMP factor homologues in Propionibacterium acnes: A new protein family differentially expressed by types I and II. Microbiology 2005, 151, 1369–1379. [Google Scholar] [CrossRef]
- Lodes, M.J.; Secrist, H.; Benson, D.R.; Jen, S.; Shanebeck, K.D.; Guderian, J.; Maisonneuve, J.F.; Bhatia, A.; Persing, D.; Patrick, S.; et al. Variable expression of immunoreactive surface proteins of Propionibacterium acnes. Microbiology 2006, 152, 3667–3681. [Google Scholar] [CrossRef]
- McDowell, A.; Gao, A.; Barnard, E.; Fink, C.; Murray, P.I.; Dowson, C.G.; Nagy, I.; Lambert, P.A.; Patrick, S. A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology 2011, 157, 1990–2003. [Google Scholar] [CrossRef]
- Alexeyev, O.A.; Dekio, I.; Layton, A.M.; Li, H.; Hughes, H.; Morris, T.; Zouboulis, C.C.; Patrick, S. Why we continue to use the name Propionibacterium acnes. Br. J. Dermatol. 2018, 179, 1227. [Google Scholar] [CrossRef]
- Dekio, I.; McDowell, A.; Sakamoto, M.; Tomida, S.; Ohkuma, M. Proposal of new combination, Cutibacterium acnes subsp. elongatum comb. nov., and emended descriptions of the genus Cutibacterium, Cutibacterium acnes subsp. acnes and Cutibacterium acnes subsp. defendens. Int. J. Syst. Evol. Microbiol. 2019, 4, 1087–1092. [Google Scholar] [CrossRef]
- Hay, R.J.; Johns, N.E.; Williams, H.C.; Bolliger, I.W.; Dellavalle, R.P.; Margolis, D.J.; Marks, R.; Naldi, L.; Weinstock, M.A.; Wulf, S.K.; et al. The global burden of skin disease in 2010: An analysis of the prevalence and impact of skin conditions. J. Investig. Dermatol. 2014, 134, 1527–1534. [Google Scholar] [CrossRef]
- Moradi Tuchayi, S.; Makrantonaki, E.; Ganceviciene, R.; Dessinioti, C.; Feldman, S.R.; Zouboulis, C.C. Acne vulgaris. Nat. Rev. Dis. Primers 2015, 1, 15029. [Google Scholar] [CrossRef]
- Tan, A.U.; Schlosser, B.J.; Paller, A.S. A review of diagnosis and treatment of acne in adult female patients. Int. J. Womens Dermatol. 2017, 4, 56–71. [Google Scholar] [CrossRef]
- Dréno, B. What is new in the pathophysiology of acne, an overview. J. Eur. Acad. Dermatol. Venereol. 2017, 31 (Suppl. 5), 8–12. [Google Scholar]
- Tanghetti, E.A. The role of inflammation in the pathology of acne. J. Clin. Aesthet. Dermatol. 2013, 9, 27–35. [Google Scholar]
- Eady, E.A.; Cove, J.H.; Holland, K.T.; Cunliffe, W.J. Erythromycin resistant propionibacteria in antibiotic-treated acne patients: Association with therapeutic failure. Br. J. Dermatol. 1989, 121, 51–57. [Google Scholar] [CrossRef]
- Puhvel, S.M.; Barfatani, M.; Warnick, M.; Sternberg, T.H. Study of antibody levels to Corynebacterium acnes in the serum of patients with acne vulgaris, using bacterial agglutination, agar gel immunodiffusion, and immunofluorescence techniques. Arch. Dermatol. 1964, 90, 421–427. [Google Scholar] [CrossRef]
- Gilchrist, T.C. A bacteriological and microscopical study of over 300 vesicular and pustular lesions of the skin, with a research upon the etiology of acne vulgaris. Johns Hopkins Hospt. Rept. 1900, 9, 409–430. [Google Scholar]
- Leyden, J.J.; McGinley, K.J.; Mills, O.; Kligman, A.M. Propionibacterium levels in patients with and without acne vulgaris. J. Investig. Dermatol. 1975, 65, 382–384. [Google Scholar] [CrossRef]
- Jahns, A.C.; Lundskog, B.; Ganceviciene, R.; Palmer, R.H.; Goloveva, I.; Zouboulis, C.C.; McDowell, A.; Patrick, S.; Alexeyev, O.A. An increased incidence of Propionibacterium acnes biofilms in acne vulgaris: A case–control study. Br. J. Dermatol. 2012, 167, 50–58. [Google Scholar] [CrossRef]
- Jahns, A.C.; Eilers, H.; Ganceviciene, R.; Alexeyev, O.A. Propionibacterium species and follicular keratinocyte activation in acneic and normal skin. Br. J. Dermatol. 2015, 172, 981–987. [Google Scholar] [CrossRef]
- Jeremy, A.H.T.; Holland, D.B.; Roberts, S.G.; Thomson, K.F.; Cunliffe, W.J. Inflammatory events are involved in acne lesion initiation. J. Investig. Dermatol. 2003, 121, 20–27. [Google Scholar] [CrossRef]
- Kligman, A.M. An overview of acne. J. Investig. Dermatol. 1974, 62, 268–287. [Google Scholar] [CrossRef]
- Ingham, E.; Eady, E.A.; Goodwin, C.E.; Cove, J.H.; Cunliffe, W.J. Pro-inflammatory levels of interleukin-1α-like bioactivity are present in the majority of open comedones in acne vulgaris. J. Investig. Dermatol. 1992, 98, 895–901. [Google Scholar] [CrossRef]
- Kelhälä, H.L.; Palatsi, R.; Fyhrquist, N.; Lehtimäki, S.; Väyrynen, J.P.; Kallioinen, M.; Kubin, M.E.; Greco, D.; Tasanen, K.; Alenius, H.; et al. Il-17/Th17 pathway is activated in acne lesions. PLoS ONE 2014, 9, e105238. [Google Scholar] [CrossRef]
- Shaheen, B.; Gonzalez, M. A microbial aetiology of acne: What is the evidence? Br. J. Dermatol. 2011, 165, 474–485. [Google Scholar] [CrossRef]
- Yu, Y.; Champer, J.; Agak, G.W.; Kao, S.; Modlin, R.L.; Kim, J. Different Propionibacterium acnes phylotypes induce distinct immune responses and express unique surface and secreted proteomes. J. Investig. Dermatol. 2016, 136, 2221–2228. [Google Scholar] [CrossRef]
- Miskin, J.E.; Farrell, A.M.; Cunliffe, W.J.; Holland, K.T. Propionibacterium acnes, a resident of lipid-rich human skin, produces a 33 kDa extracellular lipase encoded by GehA. Microbiology 1997, 143, 1745–1755. [Google Scholar] [CrossRef]
- Kim, J.; Ochoa, M.T.; Krutzik, S.R.; Takeuchi, O.; Uematsu, S.; Legaspi, A.J.; Brightbill, H.D.; Holland, D.; Cunliffe, W.J.; Akira, S.; et al. Activation of toll-like receptor 2 in acne triggers inflammatory cytokine responses. J. Immunol. 2002, 169, 1535–1541. [Google Scholar] [CrossRef]
- De Koning, H.D.; Simon, A.; Zeeuwen, P.L.J.M.; Schalkwijk, J. Pattern recognition receptors in infectious skin diseases. Microbes Infect. 2012, 14, 881–893. [Google Scholar] [CrossRef]
- Lwin, S.M.; Kimber, I.; McFadden, J.P. Acne, quorum sensing and danger. Clin. Exp. Dermatol. 2014, 39, 162–167. [Google Scholar] [CrossRef]
- Quin, M.; Pirouz, A.; Kim, M.-H.; Krutzik, S.R.; Garbán, H.J.; Kim, J. Propionibacterium acnes induces IL-1β secretion via the NLRP3 inflammasome in human monocytes. J. Investig. Dermatol. 2014, 134, 381–388. [Google Scholar] [CrossRef]
- Li, Z.J.; Choi, D.K.; Sohn, K.C.; Seo, M.S.; Lee, H.E.; Lee, Y.; Seo, Y.J.; Lee, Y.H.; Shi, G.; Zouboulis, C.C.; et al. Propionibacterium acnes activates the NLRP3 inflammasome in human sebocytes. J. Investig. Dermatol. 2014, 134, 2724–2756. [Google Scholar] [CrossRef]
- Agak, G.W.; Kao, S.; Ouyang, K.; Qin, M.; Moon, D.; Butt, A.; Kim, J. Phenotype and antimicrobial activity of Th17 cells induced by Propionibacterium acnes strain associated with healthy and acne skin. J. Investig. Dermatol. 2018, 138, 316–324. [Google Scholar] [CrossRef]
- Agak, G.W.; Qin, M.; Nobe, J.; Kim, M.-H.; Krutzik, S.R.; Tristan, G.R.; Elashoff, D.; Garban, H.J.; Kim, J. Propionibacterium acnes induces an interleukin-17 response in acne vulgaris that is regulated by vitamin A and vitamin D. J. Investig. Dermatol. 2014, 134, 366–373. [Google Scholar] [CrossRef]
- Lee, S.E.; Kim, J.-M.; Jeong, S.K.; Jeon, J.E.; Yoon, H.-J.; Jeong, M.-K.; Lee, S.H. Protease-activated receptor-2 mediates the expression of inflammatory cytokines, antimicrobial peptides, and matrix metalloproteinases in keratinocytes in response to Propionibacterium acnes. Arch. Dermatol. Res. 2010, 302, 745–756. [Google Scholar] [CrossRef]
- Omer, H.; McDowell, A.; Alexeyev, O.A. Understanding the role of Propionibacterium acnes in acne vulgaris: The critical importance of skin sampling methodologies. Clin. Dermatol. 2017, 35, 118–129. [Google Scholar] [CrossRef]
- Kwon, H.H.; Yoon, J.Y.; Suh, D.H. Analysis of distribution patterns of Propionibacterium acnes phylotypes and Peptostreptococcus species from acne lesions. Br. J. Dermatol. 2013, 169, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- Sadhasivam, S.; Sinha, M.; Saini, S.; Kaur, S.P.; Gupta, T.; Sengupta, S.; Ghosh, S.; Sardana, K. Heterogeneity and antibiotic resistance in Propionibacterium acnes isolates and its therapeutic implications: Blurring the lines between commensal and pathogenic phylotypes. Dermatol. Ther. 2016, 29, 451–454. [Google Scholar] [CrossRef]
- Nakase, K.; Hayashi, N.; Akiyama, Y.; Aoki, S.; Noguchi, N. Antimicrobial susceptibility and phylogenetic analysis of Propionibacterium acnes isolated from acne patients in Japan between 2013 and 2015. J. Dermatol. 2017, 44, 1248–1254. [Google Scholar] [CrossRef]
- Lomholt, H.B.; Scholz, C.F.P.; Brüggemann, H.; Tettelin, H.; Kilian, M. A comparative study of Cutibacterium (Propionibacterium) acnes clones from acne patients and healthy controls. Anaerobe 2017, 47, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Fitz-Gibbon, S.; Tomida, S.; Chiu, B.-H.; Nguyen, L.; Du, C.; Liu, M.; Elashoff, D.; Erfe, M.C.; Loncaric, A.; Kim, J.; et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Investig. Dermatol. 2013, 133, 2152–2160. [Google Scholar] [CrossRef] [PubMed]
- Tomida, S.; Nguyen, L.; Chiu, B.H.; Liu, J.; Sodergren, E.; Weinstock, G.M.; Li, H. Pan-genome and comparative genome analyses of Propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. mBio 2014, 4, e00003. [Google Scholar] [CrossRef]
- Barnard, E.; Shi, B.; Kang, D.; Craft, N.; Li, H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci. Rep. 2016, 6, 39491. [Google Scholar] [CrossRef] [Green Version]
- Dréno, B. Systematic review of antibiotic resistance in acne: An increasing topical and oral threat. Lancet Infect. Dis. 2016, 16, e23–e33. [Google Scholar]
- Oprica, C.; Löfmark, S.; Lund, B.; Edlund, C.; Emtestam, L.; Nord, C.E. Genetic basis of resistance in Propionibacterium acnes strains isolated from diverse types of infection in different European countries. Anaerobe 2005, 11, 137–143. [Google Scholar] [CrossRef]
- Ross, J.I.; Eady, E.A.; Carnegie, E.; Cove, J.H. Detection of transposon Tn5432-mediated macrolide-lincosamide-streptogramin B (MLSB) resistance in cutaneous propionibacteria from six European cities. J. Antimicrob. Chemother. 2002, 49, 165–168. [Google Scholar] [CrossRef] [Green Version]
- Oprica, C.; Nord, C.E. European surveillance study on the antibiotic susceptibility of Propionibacterium acnes: ESCMID study group on antimicrobial resistance in anaerobic bacteria. Clin. Microbiol. Infect. 2005, 11, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Lomholt, H.B.; Kilian, M. Clonality and anatomic distribution on the skin of antibiotic resistant and sensitive Propionibacterium acnes. Acta Derm. Venereol. 2014, 94, 534–538. [Google Scholar] [CrossRef]
- Giannopoulos, L.; Papaparaskevas, J.; Refene, E.; Daikos, G.; Stavrianeas, N.; Tsakris, A. MLST typing of antimicrobial-resistant Propionibacterium acnes isolates from patients with moderate to severe acne vulgaris. Anaerobe 2015, 31, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Patrick, S.; McDowell, A.; Lee, A.; Frau, A.; Martin, U.; Gardner, E.; McLorinan, G.; Eames, N. Antisepsis of the skin before spinal surgery with povidone iodine-alcohol followed by chlorhexidine gluconate-alcohol versus povidone iodine-alcohol applied twice for the prevention of contamination of the wound by bacteria: A randomised controlled trial. Bone Jt. J. 2017, 99-B, 1354–1365. [Google Scholar] [CrossRef] [PubMed]
- Brüggemann, H.; Henne, A.; Hoster, F.; Liesegang, H.; Wiezer, A.; Strittmatter, A.; Hujer, S.; Dürre, P.; Gottschalk, G. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 2004, 305, 671–673. [Google Scholar]
- Scholz, C.F.P.; Brüggemann, H.; Lomholt, H.B.; Tettelin, H.; Kilian, M. Genome stability of Propionibacterium acnes: A comprehensive study of indels and homopolymeric tracts. Sci. Rep. 2016, 6, 20662. [Google Scholar] [CrossRef] [PubMed]
- Brzuszkiewicz, E.; Weiner, J.; Wollherr, A.; Thürmer, A.; Hüpeden, J.; Lomholt, H.B.; Kilian, M.; Gottschalk, G.; Daniel, R.; Mollenkopf, H.J.; et al. Comparative genomics and transcriptomics of Propionibacterium acnes. PLoS ONE 2011, 6, e21581. [Google Scholar] [CrossRef]
- Kasimatis, G.; Fitz-Gibbon, S.; Tomida, S.; Wong, M.; Li, H. Analysis of complete genomes of Propionibacterium acnes reveals a novel plasmid and increased pseudogenes in an acne-associated strain. Biomed. Res. Int. 2013, 2013, 918320. [Google Scholar] [CrossRef]
- Brüggemann, H.; Lomholt, H.B.; Kilian, M. The flexible gene pool of Propionibacterium acnes. Mob. Genet. Elem. 2012, 2, 145–148. [Google Scholar] [CrossRef]
- Brüggemann, H.; Lomholt, H.B.; Tettelin, H.; Kilian, M. CRISPR/cas Loci of type II Propionibacterium acnes confer immunity against acquisition of mobile elements present in type I. P. acnes. PLoS ONE 2012, 7, e34171. [Google Scholar]
- Holland, C.; Mak, T.N.; Zimny-Arndt, U.; Schmid, M.; Meyer, T.F.; Jungblut, P.R.; Brüggemann, H. Proteomic identification of secreted proteins of Propionibacterium acnes. BMC Microbiol. 2010, 10, 230. [Google Scholar] [CrossRef]
- Lin, Y.F.; David Romero, A.; Guan, S.; Mamanova, L.; McDowall, K.J. A combination of improved differential and global RNA-seq reveals pervasive transcription initiation and events in all stages of the life-cycle of functional RNAs in Propionibacterium acnes, a major contributor to wide-spread human disease. BMC Genom. 2013, 14, 620. [Google Scholar] [CrossRef] [PubMed]
- Dekio, I.; Culak, R.; Fang, M.; Ball, G.; Gharbia, S.; Shah, H.N. Correlation between phylogroups and intracellular proteomes of Propionibacterium acnes and differences in the protein expression profiles between anaerobically and aerobically grown cells. Biomed. Res. Int. 2013, 2013, 151797. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, P.; Pot, B.; Gilluis, M.; De Vos, P.; Kersters, K.; Swings, J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996, 60, 407–438. [Google Scholar] [PubMed]
- Jeon, J.; Mok, H.J.; Choi, Y.; Park, S.C.; Jo, H.; Her, J.; Han, J.K.; Kim, Y.K.; Kim, K.P.; Ban, C. Proteomic analysis of extracellular vesicles derived from Propionibacterium acnes. Proteom. Clin. Appl. 2017, 11, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Lheure, C.; Grange, P.A.; Ollagnier, G.; Morand, P.; Désiré, N.; Sayon, S.; Corvec, S.; Raingeaud, J.; Marcelin, A.G.; Calvez, V. TLR-2 recognizes Propionibacterium acnes CAMP factor 1 from highly inflammatory strains. PLoS ONE 2016, 11, e0167237. [Google Scholar] [CrossRef] [PubMed]
- Nazipi, S.; Stødkilde, K.; Scavenius, C.; Brüggemann, H. The skin bacterium Propionibacterium acnes employs two variants of hyaluronate lyase with distinct properties. Microorganisms 2017, 5, 57. [Google Scholar] [CrossRef] [PubMed]
- Scheibner, K.A.; Lutz, M.A.; Boodoo, S.; Fenton, M.J.; Powell, J.D.; Horton, M.R. Hyaluronan fragments act as an endogenous danger signal by engaging TLR2. J. Immunol. 2006, 177, 1272–1281. [Google Scholar] [CrossRef] [PubMed]
- Termeer, C.; Benedix, F.; Sleeman, J.; Fieber, C.; Voith, U.; Ahrens, T.; Miyake, K.; Freudenberg, M.; Galanos, C.; Simon, J.C. Oligosaccharides of hyaluronan activate dendritic cells via toll-like receptor 4. J. Exp. Med. 2002, 195, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Patrick, S.; McDowell, A.; Genus, I. Propionibacterium. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Springer: New York, NY, USA, 2011; Volume 5, pp. 1138–1156. ISBN 978-0-387-95043-3. [Google Scholar]
- Von Nicolai, H.; Höffler, U.; Zilliken, F. Isolation, purification, and properties of neuraminidase from Propionibacterium acnes. Zent. Bakteriol. A 1980, 247, 84–94. [Google Scholar] [CrossRef]
- Schaller, M.; Loewenstein, M.; Borelli, C.; Jacob, K.; Vogeser, M.; Burgdorf, W.H.; Plewig, G. Induction of a chemoattractive proinflammatory cytokine response after stimulation of keratinocytes with Propionibacterium acnes and coproporphyrin III. Br. J. Dermatol. 2005, 153, 66–71. [Google Scholar] [CrossRef]
- Borelli, C.; Merk, K.; Schaller, M.; Jacob, K.; Vogeser, M.; Weindl, G.; Berger, U.; Plewig, G. In vivo porphyrin production by P. acnes in untreated acne patients and its modulation by acne treatment. Acta Derm. Venereol. 2006, 86, 316–319. [Google Scholar] [CrossRef]
- Johnson, T.; Kang, D.; Barnard, E.; Li, H. Strain-level differences in porphyrin production and regulation in Propionibacterium acnes elucidate disease associations. mSphere 2016, 1, e00023-15. [Google Scholar] [CrossRef]
- Jasson, F.; Nagy, I.; Knol, A.C.; Zuliani, T.; Khammari, A.; Dréno, B. Different strains of Propionibacterium acnes modulate differently the cutaneous innate immunity. Exp. Dermatol. 2013, 9, 587–592. [Google Scholar] [CrossRef]
- Kang, D.; Shi, B.; Erfe, M.C.; Craft, N.; Li, H. Vitamin B12 modulates the transcriptome of the skin microbiota in acne pathogenesis. Sci. Transl. Med. 2015, 293, 293ra103. [Google Scholar] [CrossRef]
- Bek-Thomsen, M.; Lomholt, H.B.; Scavenius, C.; Enghild, J.J.; Brüggemann, H. Proteome analysis of human sebaceous follicle infundibula extracted from healthy and acne-affected skin. PLoS ONE 2014, 9, e107908. [Google Scholar] [CrossRef]
- Liu, J.; Yan, R.; Zhong, Q.; Ngo, S.; Bangayan, N.J.; Nguyen, L.; Lui, T.; Liu, M.; Erfe, M.C.; Craft, N.; et al. The diversity and host interactions of Propionibacterium acnes bacteriophages on human skin. ISME J. 2015, 9, 2078–2093. [Google Scholar] [CrossRef]
- Lood, R.; Mörgelin, M.; Holmberg, A.; Rasmussen, M.; Collin, M. Inducible Siphoviruses in superficial and deep tissue isolates of Propionibacterium acnes. BMC Microbiol. 2008, 8, 139. [Google Scholar] [CrossRef]
- Marinelli, L.J.; Fitz-Gibbon, S.; Hayes, C.; Bowman, C.; Inkeles, M.; Loncaric, A.; Russell, D.A.; Jacobs-Sera, D.; Cokus, S.; Pellegrini, M.; et al. Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates. MBio 2012, 3, e00279-12. [Google Scholar]
- Brüggemann, H.; Lood, R. Bacteriophages infecting Propionibacterium acnes. Biomed. Res. Int. 2013, 2013, 705741. [Google Scholar] [CrossRef]
- Jończyk-Matysiak, E.; Weber-Dąbrowska, B.; Żaczek, M.; Międzybrodzki, R.; Letkiewicz, S.; Łusiak-Szelchowska, M.; Górski, A. Prospects of phage application in the treatment of acne caused by Propionibacterium acnes. Front. Microbiol. 2017, 8, 164. [Google Scholar] [CrossRef]
- Castillo, D.E.; Nanda, S.; Keri, J.E. Propionibacterium (Cutibacterium) acnes bacteriophage therapy in acne: Current evidence and future perspectives. Dermatol. Ther. (Heidelb) 2019, 9, 19–31. [Google Scholar] [CrossRef]
- Webster, G.F.; Cummins, C.S. Use of bacteriophage typing to distinguish Propionibacterium acnes types I and II. J. Clin. Microbiol. 1978, 7, 84–90. [Google Scholar]
- Brown, T.L.; Petrovski, S.; Dyson, Z.A.; Seviour, R.; Tucci, J. The formulation of bacteriophage in a semi solid preparation for control of Propionibacterium acnes growth. PLoS ONE 2016, 11, e0151184. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef]
- Mottin, V.H.M.; Suyenaga, E.S. An approach on the potential use of probiotics in the treatment of skin conditions: Acne and atopic dermatitis. Int. J. Dermatol. 2018, 57, 1425–1432. [Google Scholar] [CrossRef]
- Wang, Y.; Kuo, S.; Shu, M.; Yu, J.; Huang, S.; Dai, A.; Two, A.; Gallo, R.L.; Huang, C.-M. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: Implications of probiotics in acne vulgaris. Appl. Microbiol. Biotechnol. 2014, 98, 411–424. [Google Scholar] [CrossRef]
- Wang, Y.; Kao, M.S.; Yu, J.; Huang, S.; Marito, S.; Gallo, R.L.; Huang, C.-M. A precision microbiome approach using sucrose for selective augmentation of Staphylococcus epidermidis fermentation against Propionibacterium acnes. Int. J. Mol. Sci. 2016, 17, 1870. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Tang, D.C.; Zhang, L.; Gallo, R.L.; Huang, C.-M. Propionibacterium acnes CAMP factor and host acid sphingomyelinase contribute to bacterial virulence: Potential targets for inflammatory acne treatment. PLoS ONE 2011, 6, e14797. [Google Scholar] [CrossRef]
- Liu, P.F.; Nakatsuji, T.; Zhu, W.; Gallo, R.L.; Huang, C.-M. Passive immunoprotection targeting a secreted CAMP factor of Propionibacterium acnes as a novel immunotherapeutic for acne vulgaris. Vaccine 2011, 29, 3230–3238. [Google Scholar] [CrossRef]
- Wang, Y.; Hata, T.R.; Tong, Y.L.; Kao, M.S.; Zouboulis, C.C.; Gallo, R.L.; Huang, C.-M. The anti-inflammatory activities of Propionibacterium acnes CAMP factor-targeted acne vaccines. J. Investig. Dermatol. 2018, 138, 2355–2364. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Liu, Y.T.; Huang, C.P.; Zouboulis, C.C.; Gallo, R.L.; Huang, C.-M. Vaccination targeting a surface sialidase of P. acnes: Implication for new treatment of acne vulgaris. PLoS ONE 2008, 3, e1551. [Google Scholar] [CrossRef]
- Hill, C. Virulence or niche factors: what’s in a name? J. Bacteriol. 2012, 194, 5725–5727. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
| Ribotype | Type | MLST8 Clonal Complexes (CC) | N * | Percentage of Clones from Acne | Percentage of Clones from Controls | p-Value |
|---|---|---|---|---|---|---|
| RT1 | IA1/IB | CC1; CC3; CC4; CC5 | 90 | 48% | 52% | 0.84 |
| RT2 | II | CC6; CC72 | 48 | 51% | 49% | 0.36 |
| RT3 | IA2 | CC2 | 60 | 40% | 60% | 0.092 |
| RT4 | IA1 | CC3 | 23 | 84% | 16% | 0.049 |
| RT5 | IA1/IC | CC1; CC3; CC107 | 15 | 99% | 1% | 0.0005 |
| RT6 | II | CC6 | 11 | 1% | 99% | 0.02 |
| RT7 | ND | ND | 10 | 99% | 1% | 0.12 |
| RT8 | IA1 | CC4 | 5 | 100% | 0% | 0.024 |
| RT9 | III | CC77 | 4 | 99% | 1% | 0.29 |
| RT10 | ND | ND | 5 | 100% | 0% | 0.024 |
| Putative Gene Function | KPA171202 ORF Number | Potential Effect on the Host |
|---|---|---|
| CAMP-factors | PPA0687, PPA1198, PPA1231, PPA1340, PPA2108 | Haemolytic, cytotoxic |
| Haemolysins | PPA0565, PPA0938, PPA1396 | Haemolytic |
| GehA lipases | PPA1796, PPA2105 | Tissue damage, inflammation |
| Sialidases | PPA0684, PPA0685, PPA1560 | Tissue damage |
| Hyaluronate lyase (HYL) | PPA0380 | Tissue damage |
| Endoglycoceramidases | PPA0644, PPA2106 | Tissue damage |
| Endo-ß-N-acetylglucosaminidase | PPA0990 | Tissue damage |
| Dermatan sulphate adhesin (DsA1) (PTR) * | PPA2127 | Colonization/adhesion/Fibrinogen-binding/inflammation |
| Dermatan sulphate adhesin (DsA2) (PTR) * | PPA2210 | Colonization/adhesion/inflammation |
| PTRs * | PPA1715, PPA1879, PPA180, PPA1881, PPA1906, PPA2130, PPA2270 | Colonization/adhesion/inflammation |
| GroEL | PPA0453, PPA1772, PPA1773 | Inflammation |
| DnaK | PPA1098, PPA2040 | Inflammation |
| DnaJ | PPA0916, PPA2038 | Inflammation |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McLaughlin, J.; Watterson, S.; Layton, A.M.; Bjourson, A.J.; Barnard, E.; McDowell, A. Propionibacterium acnes and Acne Vulgaris: New Insights from the Integration of Population Genetic, Multi-Omic, Biochemical and Host-Microbe Studies. Microorganisms 2019, 7, 128. https://doi.org/10.3390/microorganisms7050128
McLaughlin J, Watterson S, Layton AM, Bjourson AJ, Barnard E, McDowell A. Propionibacterium acnes and Acne Vulgaris: New Insights from the Integration of Population Genetic, Multi-Omic, Biochemical and Host-Microbe Studies. Microorganisms. 2019; 7(5):128. https://doi.org/10.3390/microorganisms7050128
Chicago/Turabian StyleMcLaughlin, Joseph, Steven Watterson, Alison M. Layton, Anthony J. Bjourson, Emma Barnard, and Andrew McDowell. 2019. "Propionibacterium acnes and Acne Vulgaris: New Insights from the Integration of Population Genetic, Multi-Omic, Biochemical and Host-Microbe Studies" Microorganisms 7, no. 5: 128. https://doi.org/10.3390/microorganisms7050128





