Polyionic Tags as Enhancers of Protein Solubility in Recombinant Protein Expression
Abstract
:1. Introduction
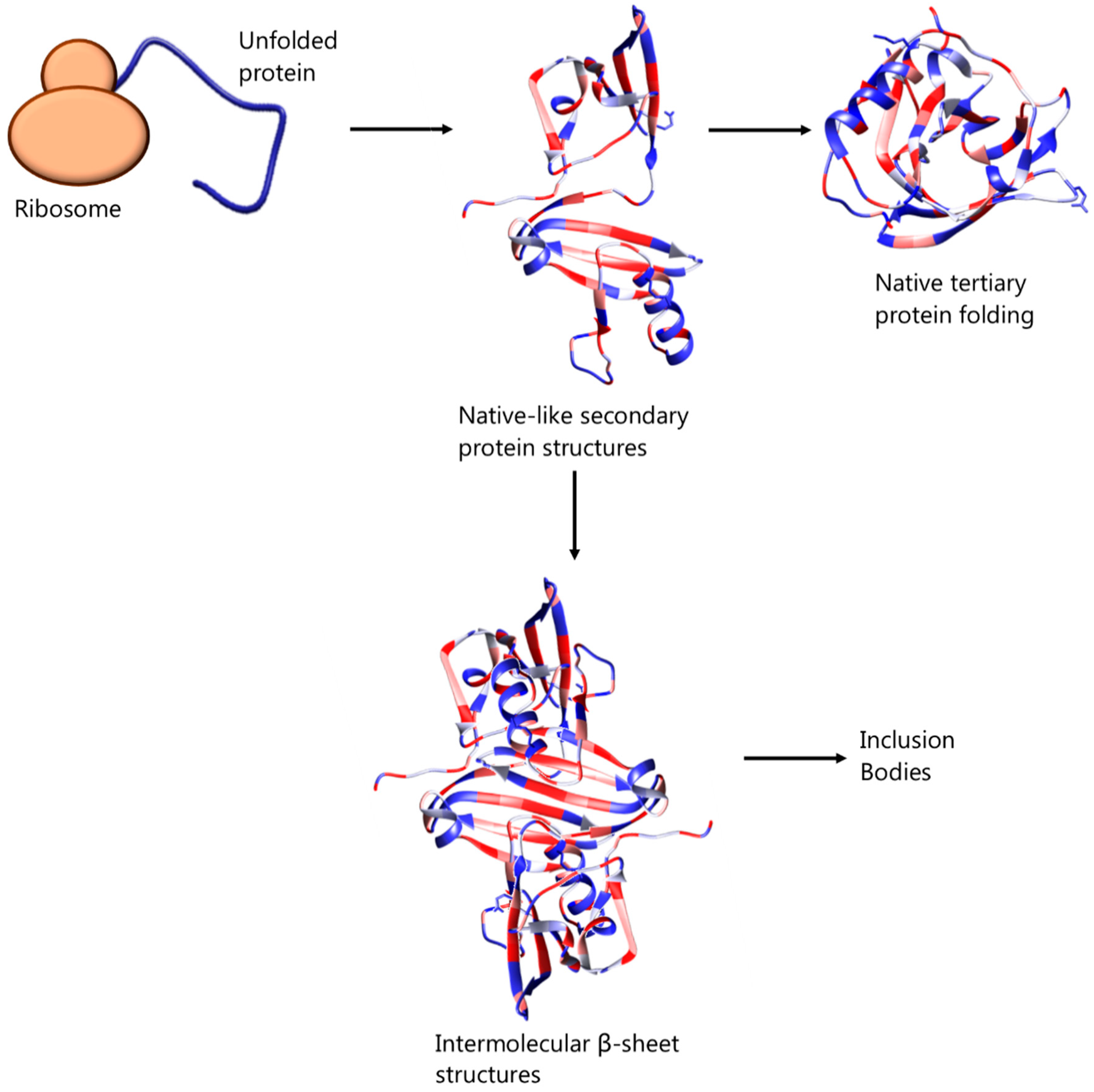
2. Inclusion Bodies and Their Avoidance
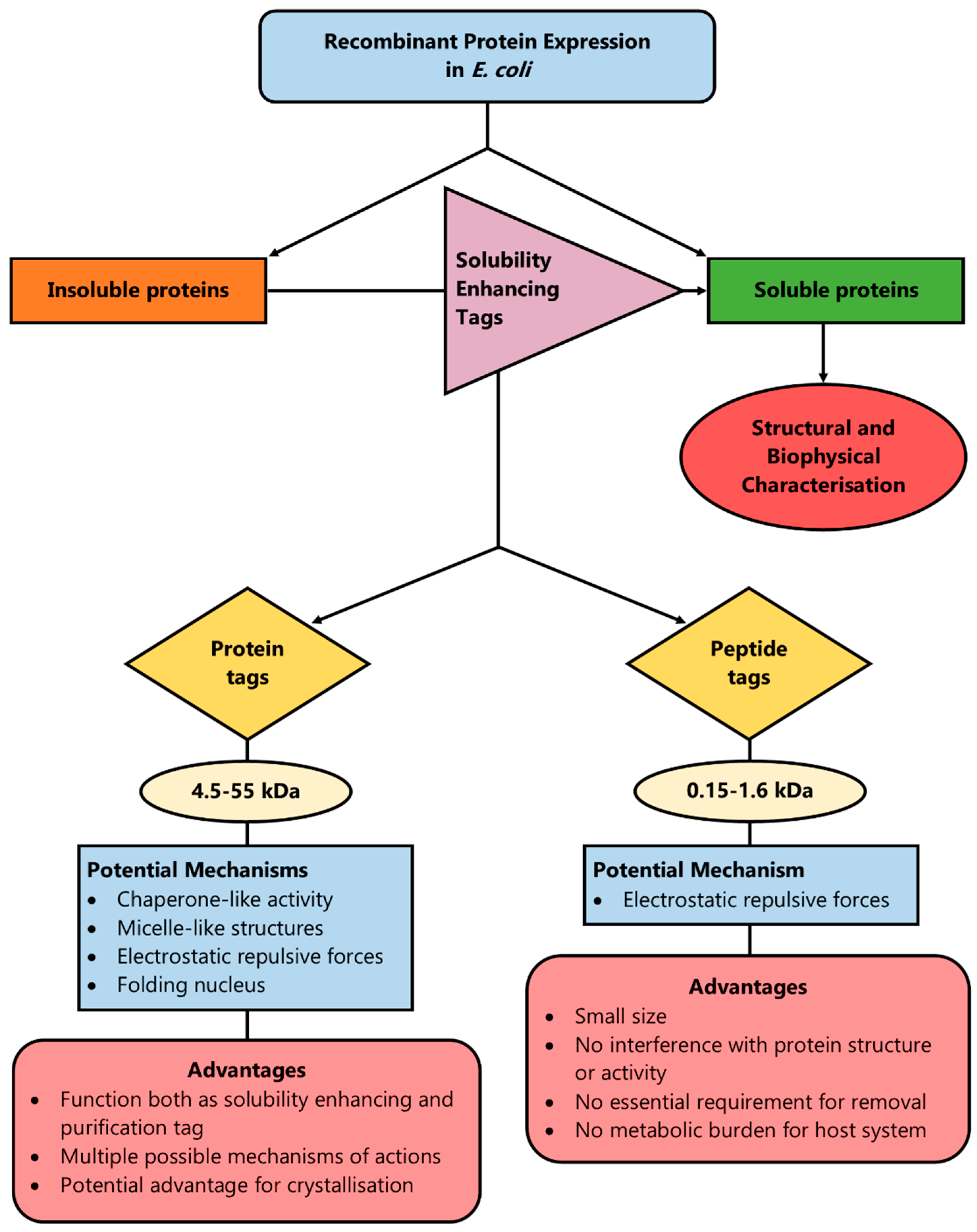
3. Protein Fusion Tags
4. Peptide Tags
4.1. Polycationic Tags as Enhancers of Protein Solubility in Recombinant Protein Production
4.2. Solubilising Peptide Tags in Solid-Phase Peptide Synthesis (SPPS) and Native Chemical Ligation
4.3. Polyanionic Tags as Enhancers of Protein Solubility in Recombinant Protein Production
4.4. Polycationic versus Polyanionic Tags
4.5. Polyionic Tags Displaying the Opposite Effect
5. Supercharging of Proteins
6. Discussion
7. Conclusions
Acknowledgments
Conflicts of Interest
References
- Itakura, K.; Hirose, T.; Crea, R.; Riggs, A.D.; Heyneker, H.L.; Bolivar, F.; Boyer, H.W. Expression in Escherichia coli of a chemically synthesized gene for the hormone somatostatin. Science 1977, 198, 1056–1063. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, S.A.; Engen, J.R.; Mazzeo, J.R.; Jones, G.B. Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nat. Rev. Drug Discov. 2012, 11, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L.; Vaishnav, P. Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 2009, 27, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Grangeasse, C.; Stülke, J.; Mijakovic, I. Regulatory potential of post-translational modifications in bacteria. Front. Microbiol. 2015, 6, 500. [Google Scholar] [CrossRef] [PubMed]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli Physiology in Luria-Bertani Broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, H.P.; Mortensen, K.K. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 2005, 115, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Mitraki, A.; Haase-Pettingell, C.; King, J. Mechanisms of Inclusion Body Formation. In Protein Refolding; ACS Publications: Washington, DC, USA, 1991; pp. 35–49. [Google Scholar]
- Ami, D.; Natalello, A.; Gatti-Lafranconi, P.; Lotti, M.; Doglia, S.M. Kinetics of inclusion body formation studied in intact cells by FT-IR spectroscopy. FEBS Lett. 2005, 579, 3433–3436. [Google Scholar] [CrossRef] [PubMed]
- Carrio, M.; Gonzalez-Montalban, N.; Vera, A.; Villaverde, A.; Ventura, S. Amyloid-like properties of bacterial inclusion bodies. J. Mol. Biol. 2005, 347, 1025–1037. [Google Scholar] [CrossRef] [PubMed]
- Ami, D.; Natalello, A.; Taylor, G.; Tonon, G.; Doglia, S.M. Structural analysis of protein inclusion bodies by Fourier transform infrared microspectroscopy. Biochim. Biophys. Acta Proteins Proteom. 2006, 1764, 793–799. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Maji, S.K.; Sawaya, M.R.; Eisenberg, D.; Riek, R. Bacterial Inclusion Bodies Contain Amyloid-Like Structure. PLoS Biol. 2008, 6, e195. [Google Scholar] [CrossRef] [PubMed]
- Fink, A.L. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Fold. Des. 1998, 3, R9–R23. [Google Scholar] [CrossRef]
- Strandberg, L.; Enfors, S.O. Factors influencing inclusion body formation in the production of a fused protein in Escherichia coli. Appl. Environ. Microbiol. 1991, 57, 1669–1674. [Google Scholar] [PubMed]
- Rinas, U.; Garcia-Fruitos, E.; Corchero, J.L.; Vazquez, E.; Seras-Franzoso, J.; Villaverde, A. Bacterial Inclusion Bodies: Discovering Their Better Half. Trends Biochem. Sci. 2017, 42, 726–737. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fruitos, E.; Vazquez, E.; Diez-Gil, C.; Corchero, J.L.; Seras-Franzoso, J.; Ratera, I.; Veciana, J.; Villaverde, A. Bacterial inclusion bodies: Making gold from waste. Trends Biotechnol. 2012, 30, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Middelberg, A.P.J. Preparative protein refolding. Trends Biotechnol. 2002, 20, 437–443. [Google Scholar] [CrossRef]
- Cabrita, L.D.; Bottomley, S.P. Protein expression and refolding—A practical guide to getting the most out of inclusion bodies. Biotechnol. Annu. Rev. 2004, 10, 31–50. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, G.; Valax, P.; Ostermeier, M.; Horowitz, P.M. Folding and aggregation of TEM beta-lactamase: Analogies with the formation of inclusion bodies in Escherichia coli. Protein Sci. 1994, 3, 1953–1960. [Google Scholar] [CrossRef] [PubMed]
- Caspers, P.; Stieger, M.; Burn, P. Overproduction of bacterial chaperones improves the solubility of recombinant protein tyrosine kinases in Escherichia coli. Cell. Mol. Biol. 1994, 40, 635–644. [Google Scholar] [PubMed]
- Cole, P.A. Chaperone-assisted protein expression. Structure 1996, 4, 239–242. [Google Scholar] [CrossRef]
- Makrides, S.C. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 1996, 60, 512–538. [Google Scholar] [PubMed]
- Derman, A.I.; Prinz, W.A.; Belin, D.; Beckwith, J. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science 1993, 262, 1744–1747. [Google Scholar] [CrossRef] [PubMed]
- Esposito, D.; Chatterjee, D.K. Enhancement of soluble protein expression through the use of fusion tags. Curr. Opin. Biotechnol. 2006, 17, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.-J.; Kim, S.-K.; Min, W.-K.; Lee, S.-S.; Park, K.; Park, Y.-C.; Seo, J.-H. Polycationic amino acid tags enhance soluble expression of Candida antarctica lipase B in recombinant Escherichia coli. Bioprocess Biosyst. Eng. 2011, 34, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-G.; Min, W.-K.; Rho, Y.-T.; Seo, J.-H. Electrostatic interaction-induced inclusion body formation of glucagon-like peptide-1 fused with ubiquitin and cationic tag. Protein Expr. Purif. 2012, 84, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Hage, N.; Renshaw, J.G.; Winkler, G.S.; Gellert, P.; Stolnik, S.; Falcone, F.H. Improved expression and purification of the Helicobacter pylori adhesin BabA through the incorporation of a hexa-lysine tag. Protein Expr. Purif. 2015, 106, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.J.; Almeida, A.; Castro, A.; Domingues, L.; Besir, H. The novel Fh8 and H fusion partners for soluble protein expression in Escherichia coli: A comparison with the traditional gene fusion technology. Appl. Microbiol. Biotechnol. 2013, 97, 6779–6791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.B.; Johnson, K.S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 1988, 67, 31–40. [Google Scholar] [CrossRef]
- LaVallie, E.R.; DiBlasio, E.A.; Kovacic, S.; Grant, K.L.; Schendel, P.F.; McCoy, J.M. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology 1993, 11, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Maina, C.V.; Riggs, P.D.; Grandea, A.G.; Slatko, B.E.; Moran, L.S.; Tagliamonte, J.A.; McReynolds, L.A.; di Guan, C. An Escherichia coli vector to express and purify foreign proteins by fusion to and separation from maltose-binding protein. Gene 1988, 74, 365–373. [Google Scholar] [CrossRef]
- Kapust, R.B.; Waugh, D.S. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999, 8, 1668–1674. [Google Scholar] [CrossRef] [PubMed]
- Butt, T.R.; Jonnalagadda, S.; Monia, B.P.; Sternberg, E.J.; Marsh, J.A.; Stadel, J.M.; Ecker, D.J.; Crooke, S.T. Ubiquitin fusion augments the yield of cloned gene products in Escherichia coli. Proc. Natl. Acad. Sci. USA 1989, 86, 2540–2544. [Google Scholar] [CrossRef] [PubMed]
- Samuelsson, E.; Moks, T.; Uhlen, M.; Nilsson, B. Enhanced in vitro Refolding of Insulin-like Growth Factor I Using a Solubilizing Fusion Partner. Biochemistry 1994, 33, 4207–4211. [Google Scholar] [CrossRef] [PubMed]
- Huth, J.R.; Bewley, C.A.; Jackson, B.M.; Hinnebusch, A.G.; Clore, G.M.; Gronenborn, A.M. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 1997, 6, 2359–2364. [Google Scholar] [CrossRef] [PubMed]
- Collins-Racie, L.A.; McColgan, J.M.; Grant, K.L.; DiBlasio-Smith, E.A.; McCoy, J.M.; LaVallie, E.R. Production of recombinant bovine enterokinase catalytic subunit in Escherichia coli using the novel secretory fusion partner DsbA. Biotechnology 1995, 13, 982–987. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Olsen, D.R.; Nguyen, K.B.; Olson, P.S.; Rhodes, E.T.; Mascarenhas, D. Expression of Eukaryotic Proteins in Soluble Form in Escherichia coli. Protein Expr. Purif. 1998, 12, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.D.; Elisee, C.; Newham, D.M.; Harrison, R.G. New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol. Bioeng. 1999, 65, 382–388. [Google Scholar] [CrossRef]
- Sorensen, H.P.; Sperling-Petersen, H.U.; Mortensen, K.K. A favorable solubility partner for the recombinant expression of streptavidin. Protein Expr. Purif. 2003, 32, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Reddi, H.; Bhattacharya, A.; Kumar, V. The calcium-binding protein of Entamoeba histolytica as a fusion partner for expression of peptides in Escherichia coli. Biotechnol. Appl. Biochem. 2002, 36, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Malakhov, M.P.; Mattern, M.R.; Malakhova, O.A.; Drinker, M.; Weeks, S.D.; Butt, T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genom. 2004, 5, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.-Y.; Choi, H.; Kim, Y.-H.; Han, K.-Y.; Park, J.-S.; Han, S.-S.; Lee, J. Heterologous gene expression using self-assembled supra-molecules with high affinity for HSP70 chaperone. Nucleic Acids Res. 2005, 33, 3751–3762. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, D.K.; Esposito, D. Enhanced soluble protein expression using two new fusion tags. Protein Expr. Purif. 2006, 46, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Rudolph, R.; Söhling, B. A novel fusion protein system for the production of native human pepsinogen in the bacterial periplasm. Protein Expr. Purif. 2006, 47, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K.-Y.; Song, J.-A.; Han, K.-Y.; Park, J.-S.; Seo, H.-S.; Lee, J. Heterologous protein expression using a novel stress-responsive protein of E. coli RpoA as fusion expression partner. Enzyme Microb. Technol. 2007, 41, 859–866. [Google Scholar] [CrossRef]
- Han, K.-Y.; Seo, H.-S.; Song, J.-A.; Ahn, K.-Y.; Park, J.-S.; Lee, J. Transport proteins PotD and Crr of Escherichia coli, novel fusion partners for heterologous protein expression. Biochim. Biophys. Acta 2007, 1774, 1536–1543. [Google Scholar] [CrossRef] [PubMed]
- Han, K.-Y.; Song, J.-A.; Ahn, K.-Y.; Park, J.-S.; Seo, H.-S.; Lee, J. Enhanced solubility of heterologous proteins by fusion expression using stress-induced Escherichia coli protein, Tsf. FEMS Microbiol. Lett. 2007, 274, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Han, K.-Y.; Song, J.-A.; Ahn, K.-Y.; Park, J.-S.; Seo, H.-S.; Lee, J. Solubilization of aggregation-prone heterologous proteins by covalent fusion of stress-responsive Escherichia coli protein, SlyD. Protein Eng. Des. Sel. 2007, 20, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Zou, Z.; Feng, S.; Zhou, P.; Cao, L. The acidity of protein fusion partners predominantly determines the efficacy to improve the solubility of the target proteins expressed in Escherichia coli. J. Biotechnol. 2007, 129, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-S.; Han, K.-Y.; Lee, J.-H.; Song, J.-A.; Ahn, K.-Y.; Seo, H.-S.; Sim, S.-J.J.; Kim, S.-W.; Lee, J. Solubility enhancement of aggregation-prone heterologous proteins by fusion expression using stress-responsive Escherichia coli protein, RpoS. BMC Biotechnol. 2008, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Cao, L.; Zhou, P.; Su, Y.; Sun, Y.; Li, W. Hyper-acidic protein fusion partners improve solubility and assist correct folding of recombinant proteins expressed in Escherichia coli. J. Biotechnol. 2008, 135, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Ohana, R.F.; Encell, L.P.; Zhao, K.; Simpson, D.; Slater, M.R.; Urh, M.; Wood, K. V HaloTag7: A genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification. Protein Expr. Purif. 2009, 68, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wu, D.; Lu, Z.; Chen, W.; Hu, X.; Ding, Y. A Novel Method for High-Level Production of TEV Protease by Superfolder GFP Tag. J. Biomed. Biotechnol. 2009, 2009. [Google Scholar] [CrossRef] [PubMed]
- DelProposto, J.; Majmudar, C.Y.; Smith, J.L.; Brown, W.C. Mocr: A novel fusion tag for enhancing solubility that is compatible with structural biology applications. Protein Expr. Purif. 2009, 63, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Caswell, J.; Snoddy, P.; McMeel, D.; Buick, R.J.; Scott, C.J. Production of recombinant proteins in Escherichia coli using an N-terminal tag derived from sortase. Protein Expr. Purif. 2010, 70, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Gu, J.; Wang, H.; Yu, S.; Liu, Y.; Ning, Y.; Zou, Q.; Yu, X.; Mao, X. EspA is a novel fusion partner for expression of foreign proteins in Escherichia coli. J. Biotechnol. 2010, 150, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-A.; Lee, D.-S.; Park, J.-S.; Han, K.-Y.; Lee, J. A novel Escherichia coli solubility enhancer protein for fusion expression of aggregation-prone heterologous proteins. Enzyme Microb. Technol. 2011, 49, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, H.; Saito, S.; Sakai, K.; Yamaguchi, R.; Katsuyama, I.; Arakawa, T.; Onozaki, K.; Arakawa, T.; Tokunaga, M. Halophilic beta-lactamase as a new solubility- and folding-enhancing tag protein: Production of native human interleukin 1alpha and human neutrophil alpha-defensin. Appl. Microbiol. Biotechnol. 2010, 86, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Hansted, J.G.; Pietikainen, L.; Hog, F.; Sperling-Petersen, H.U.; Mortensen, K.K. Expressivity tag: A novel tool for increased expression in Escherichia coli. J. Biotechnol. 2011, 155, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Cortez, T.; Morones-Ramirez, J.R.; Balderas-Renteria, I.; Zarate, X. Expression and purification of recombinant proteins in Escherichia coli tagged with a small metal-binding protein from Nitrosomonas europaea. Protein Expr. Purif. 2016, 118, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wu, S.; Cui, L.; Wu, Y.; Jiang, T.; He, B. A novel Ffu fusion system for secretory expression of heterologous proteins in Escherichia coli. Microb. Cell Fact. 2017, 16, 231. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Jiang, L.; Wang, W.; Wang, R.; Fan, J. Evaluation of rice tetraticopeptide domain-containing thioredoxin as a novel solubility-enhancing fusion tag in Escherichia coli. J. Biosci. Bioeng. 2018, 125, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Guo, W.; Su, B.; Guo, Y.; Wang, J.; Chu, B.; Yang, G. High-level expression of soluble recombinant proteins in Escherichia coli using an HE-maltotriose-binding protein fusion tag. Protein Expr. Purif. 2018, 142, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Marblestone, J.G.; Edavettal, S.C.; Lim, Y.; Lim, P.; Zuo, X.; Butt, T.R. Comparison of SUMO fusion technology with traditional gene fusion systems: Enhanced expression and solubility with SUMO. Protein Sci. 2006, 15, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.; Almeida, A.; Castro, A.; Domingues, L. Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: The novel Fh8 system. Front. Microbiol. 2014, 5, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sachdev, D.; Chirgwin, J.M. Solubility of Proteins Isolated from Inclusion Bodies Is Enhanced by Fusion to Maltose-Binding Protein or Thioredoxin. Protein Expr. Purif. 1998, 12, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Maki, K.; Ebina, T.; Kuwajima, K.; Soda, K.; Kuroda, Y. Mutational analysis of protein solubility enhancement using short peptide tags. Biopolymers 2007, 85, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Smyth, D.R.; Mrozkiewicz, M.K.; McGrath, W.J.; Listwan, P.; Kobe, B. Crystal structures of fusion proteins with large-affinity tags. Protein Sci. 2003, 12, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Moon, A.F.; Mueller, G.A.; Zhong, X.; Pedersen, L.C. A synergistic approach to protein crystallization: Combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 2010, 19, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Chuenchor, W.; Jiang, J.; Cheng, J.; Li, Y.; Fang, K.; Huang, M.; Smith, P.; Xiao, T.S. Design of an expression system to enhance MBP-mediated crystallization. Sci. Rep. 2017, 7, 40991. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, E.; Venturini, S.; Pessi, A.; Tramontano, A.; Sollazzo, M. High level expression and rational mutagenesis of a designed protein, the minibody. From an insoluble to a soluble molecule. J. Mol. Biol. 1994, 236, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Hage, N.; Howard, T.; Phillips, C.; Brassington, C.; Overman, R.; Debreczeni, J.; Gellert, P.; Stolnik, S.; Winkler, G.S.; Falcone, F.H. Structural basis of Lewis(b) antigen binding by the Helicobacter pylori adhesin BabA. Sci. Adv. 2015, 1, e1500315. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Nakamura, S.; Noguchi, K.; Yohda, M.; Kidokoro, S.; Kuroda, Y. Analysis and Control of Protein Crystallization Using Short Peptide Tags That Change Solubility without Affecting Structure, Thermal Stability, and Function. Cryst. Growth Des. 2015, 15, 2703–2711. [Google Scholar] [CrossRef]
- Pessi, A.; Bianchi, E.; Crameri, A.; Venturini, S.; Tramontano, A.; Sollazzo, M. A designed metal-binding protein with a novel fold. Nature 1993, 362, 367. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wilson, S.; Elliott, T. A mutant HemA protein with positive charge close to the N terminus is stabilized against heme-regulated proteolysis in Salmonella typhimurium. J. Bacteriol. 1999, 181, 6033–6041. [Google Scholar] [PubMed]
- Park, S.H.; Mrse, A.A.; Nevzorov, A.A.; Mesleh, M.F.; Oblatt-Montal, M.; Montal, M.; Opella, S.J. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J. Mol. Biol. 2003, 333, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.C.; Derbyshire, R.B.; Cook, E.; Dunthorne, L.; Viney, J.; Brewer, S.J.; Sassenfeld, H.M.; Bell, L.D. Chemical synthesis and cloning of a poly(arginine)-coding gene fragment designed to aid polypeptide purification. Gene 1984, 32, 321–327. [Google Scholar] [CrossRef]
- Nautiyal, K.; Kuroda, Y. A SEP tag enhances the expression, solubility and yield of recombinant TEV protease without altering its activity. New Biotechnol. 2018, 42, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Woestenenk, E.A.; Hammarstrom, M.; van den Berg, S.; Hard, T.; Berglund, H. His tag effect on solubility of human proteins produced in Escherichia coli: A comparison between four expression vectors. J. Struct. Funct. Genom. 2004, 5, 217–229. [Google Scholar] [CrossRef]
- Busso, D.; Kim, R.; Kim, S.-H. Expression of soluble recombinant proteins in a cell-free system using a 96-well format. J. Biochem. Biophys. Methods 2003, 55, 233–240. [Google Scholar] [CrossRef]
- Mohanty, A.K.; Wiener, M.C. Membrane protein expression and production: Effects of polyhistidine tag length and position. Protein Expr. Purif. 2004, 33, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Hammarström, M.; Hellgren, N.; van den Berg, S.; Berglund, H.; Härd, T. Rapid screening for improved solubility of small human proteins produced as fusion proteins in Escherichia coli. Protein Sci. 2002, 11, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Porath, J.; Carlsson, J.; Olsson, I.; Belfrage, G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 1975, 258, 598–599. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.S.; Robinson, W.E. Purification and characterization of a histidine-rich glycoprotein that binds cadmium from the blood plasma of the bivalve Mytilus edulis. Arch. Biochem. Biophys. 1999, 366, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Bornhorst, J.A.; Falke, J.J. [16] Purification of Proteins Using Polyhistidine Affinity Tags. Methods Enzymol. 2000, 326, 245–254. [Google Scholar] [PubMed]
- Crowe, J.; Dobeli, H.; Gentz, R.; Hochuli, E.; Stuber, D.; Henco, K. 6xHis-Ni-NTA chromatography as a superior technique in recombinant protein expression/purification. Methods Mol. Biol. 1994, 31, 371–387. [Google Scholar] [CrossRef] [PubMed]
- Rüdiger, S.; Buchberger, A.; Bukau, B. Interaction of Hsp70 chaperones with substrates. Nat. Struct. Biol. 1997, 4, 342. [Google Scholar] [CrossRef] [PubMed]
- Hirtreiter, A.M.; Calloni, G.; Forner, F.; Scheibe, B.; Puype, M.; Vandekerckhove, J.; Mann, M.; Hartl, F.U.; Hayer-Hartl, M. Differential substrate specificity of group I and group II chaperonins in the archaeon Methanosarcina mazei. Mol. Microbiol. 2009, 74, 1152–1168. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Hidese, R.; Fujiwara, S. Function of a thermophilic archaeal chaperonin is enhanced by electrostatic interactions with its targets. J. Biosci. Bioeng. 2017, 124, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Wayne, N.; Bolon, D.N. Charge-rich regions modulate the anti-aggregation activity of Hsp90. J. Mol. Biol. 2010, 401, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Englebretsen, D.R.; Alewood, P.F. Boc SPPS of two hydrophobic peptides using a “solubilising tail” strategy: Dodecaalanine and chemotactic protein 1042-55. Tetrahedron Lett. 1996, 37, 8431–8434. [Google Scholar] [CrossRef]
- Englebretsen, D.R.; Robillard, G.T. An N-terminal method for peptide solubilisation. Tetrahedron 1999, 55, 6623–6634. [Google Scholar] [CrossRef]
- Choma, C.T.; Robillard, G.T.; Englebretsen, D.R. Synthesis of hydrophobic peptides: An Fmoc “solubilising tail” method. Tetrahedron Lett. 1998, 39, 2417–2420. [Google Scholar] [CrossRef]
- Hossain, M.A.; Belgi, A.; Lin, F.; Zhang, S.; Shabanpoor, F.; Chan, L.; Belyea, C.; Truong, H.-T.; Blair, A.R.; Andrikopoulos, S.; et al. Use of a temporary “solubilizing” peptide tag for the Fmoc solid-phase synthesis of human insulin glargine via use of regioselective disulfide bond formation. Bioconjug. Chem. 2009, 20, 1390–1396. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P.E.; Muir, T.W.; Clark-Lewis, I.; Kent, S.B. Synthesis of proteins by native chemical ligation. Science 1994, 266, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Saito, Y.; Aimoto, S. Synthesis of the C-terminal region of opioid receptor like 1 in an SDS micelle by the native chemical ligation: Effect of thiol additive and SDS concentration on ligation efficiency. J. Pept. Sci. 2005, 11, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.C.B.; Malito, E.; Shen, Y.; Rich, D.; Tang, W.-J.; Kent, S.B.H. Modular Total Chemical Synthesis of a Human Immunodeficiency Virus Type 1 Protease. J. Am. Chem. Soc. 2007, 129, 11480–11490. [Google Scholar] [CrossRef] [PubMed]
- Rathnayaka, T.; Tawa, M.; Nakamura, T.; Sohya, S.; Kuwajima, K.; Yohda, M.; Kuroda, Y. Solubilization and folding of a fully active recombinant Gaussia luciferase with native disulfide bonds by using a SEP-Tag. Biochim. Biophys. Acta 2011, 1814, 1775–1778. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-K.; Min, W.-K.; Park, Y.-C.; Seo, J.-H. Application of repeated aspartate tags to improving extracellular production of Escherichia coli L-asparaginase isozyme II. Enzyme Microb. Technol. 2015, 79–80, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-B.; Howitt, J.; McCorkle, S.; Lawrence, P.; Springer, K.; Freimuth, P. Protein aggregation during overexpression limited by peptide extensions with large net negative charge. Protein Expr. Purif. 2004, 36, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Khan, M.A.; Kuroda, Y. Analysis of amino acid contributions to protein solubility using short peptide tags fused to a simplified BPTI variant. Biochim. Biophys. Acta Proteins Proteom. 2012, 1824, 1144–1150. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Islam, M.M.; Kuroda, Y. Analysis of protein aggregation kinetics using short amino acid peptide tags. Biochim. Biophys. Acta 2013, 1834, 2107–2115. [Google Scholar] [CrossRef] [PubMed]
- Vlasuk, G.P.; Inouye, S.; Ito, H.; Itakura, K.; Inouye, M. Effects of the complete removal of basic amino acid residues from the signal peptide on secretion of lipoprotein in Escherichia coli. J. Biol. Chem. 1983, 258, 7141–7148. [Google Scholar] [PubMed]
- Kim, S.-K.; Park, Y.-C.; Lee, H.H.; Jeon, S.T.; Min, W.-K.; Seo, J.-H. Simple amino acid tags improve both expression and secretion of Candida antarctica lipase B in recombinant Escherichia coli. Biotechnol. Bioeng. 2015, 112, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Beckwith, J.; Inouye, H. Alteration of the amino terminus of the mature sequence of a periplasmic protein can severely affect protein export in Escherichia coli. Proc. Natl. Acad. Sci. USA 1988, 85, 7685–7689. [Google Scholar] [CrossRef] [PubMed]
- Sung, C.Y.; Gennity, J.M.; Pollitt, N.S.; Inouye, M. A positive residue in the hydrophobic core of the Escherichia coli lipoprotein signal peptide suppresses the secretion defect caused by an acidic amino terminus. J. Biol. Chem. 1992, 267, 997–1000. [Google Scholar] [PubMed]
- Lilie, H.; Richter, S.; Bergelt, S.; Frost, S.; Gehle, F. Polyionic and cysteine-containing fusion peptides as versatile protein tags. Biol. Chem. 2013, 394, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Stubenrauch, K.; Bachmann, A.; Rudolph, R.; Lilie, H. Purification of a viral coat protein by an engineered polyionic sequence. J. Chromatogr. B Biomed. Sci. Appl. 2000, 737, 77–84. [Google Scholar] [CrossRef]
- Richter, S.A.; Stubenrauch, K.; Lilie, H.; Rudolph, R. Polyionic fusion peptides function as specific dimerization motifs. Protein Eng. Des. Sel. 2001, 14, 775–783. [Google Scholar] [CrossRef]
- Kurinomaru, T.; Maruyama, T.; Izaki, S.; Handa, K.; Kimoto, T.; Shiraki, K. Protein-poly(amino acid) complex precipitation for high-concentration protein formulation. J. Pharm. Sci. 2014, 103, 2248–2254. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Phillips, K.J.; Liu, D.R. Supercharging Proteins Can Impart Unusual Resilience. J. Am. Chem. Soc. 2007, 129, 10110–10112. [Google Scholar] [CrossRef] [PubMed]
- Simeonov, P.; Berger-Hoffmann, R.; Hoffmann, R.; Strater, N.; Zuchner, T. Surface supercharged human enteropeptidase light chain shows improved solubility and refolding yield. Protein Eng. Des. Sel. 2011, 24, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Schein, C.H. Production of Soluble Recombinant Proteins in Bacteria. Nat. Biotechnol. 1989, 7, 1141. [Google Scholar] [CrossRef]
- Wilkinson, D.L.; Harrison, R.G. Predicting the solubility of recombinant proteins in Escherichia coli. Biotechnology 1991, 9, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.L.; Grimsley, G.R.; Yakovlev, G.I.; Makarov, A.A.; Pace, C.N. The effect of net charge on the solubility, activity, and stability of ribonuclease Sa. Protein Sci. 2001, 10, 1206–1215. [Google Scholar] [CrossRef] [PubMed]
- Hirose, S.; Kawamura, Y.; Mori, M.; Yokota, K.; Noguchi, T.; Goshima, N. Development and evaluation of data-driven designed tags (DDTs) for controlling protein solubility. New Biotechnol. 2011, 28, 225–231. [Google Scholar] [CrossRef] [PubMed]
| Name | Full Name | Size (kDa) | Reference |
|---|---|---|---|
| GST | Glutathione-S-transferase | 26 | Smith et al., 1988 [29] |
| MBP | Maltose-binding protein | 42.5 | Maina et al., 1988 [31] |
| UB | Ubiquitin | ~9 | Butt et al., 1989 [33] |
| Trx | Thioredoxin | 11.7 | LaVallie et al., 1993 [30] |
| Z-tag/ZZ-tag | IgG-binding domain from protein A | 15.5/31 | Samuelsson et al., 1994 [34] |
| GB1 | Immunoglobulin-binding domain of protein G | 6.2 | Huth et al., 1997 [35] |
| DsbA | Disulphide isomerase I | 21.1 | Collins-Racie et al., 1998 [36] |
| DsbAmut | Zhang et al., 1998 [37] | ||
| NusA | N-utilization substance A | 55 | Davis et al., 1999 [38] |
| IF2 domain I (or InfB(1-471) | Initiation factor 2 | Sorensen et al., 2003 [39] | |
| CaBP | Calcium binding protein | Reddi et al., 2002 [40] | |
| SUMO | Small ubiquitin-related modifier | 31 | Malakhov et al., 2004 [41] |
| FTN-H | Ferritin heavy-chain | Ahn et al., 2005 [42] | |
| Skp | Seventeen kilodalton protein | 17 | Chatterjee et al., 2006 [43] |
| T7PK | T7 protein kinase | 4.5 | Chatterjee et al., 2006 [43] |
| Ecotin | E. coli trypsin inhibitor | 16 | Malik et al., 2006 [44] |
| RpoA | RNA Polymerase α-subunit | 39.5 | Ahn et al., 2007 [45] |
| PotD | Spermidine/putrescine-binding periplasmic protein | 39.8 | Han et al., 2007 [46] |
| Crr | Glucose-specific phosphotransferase (PTS) enzyme IIA component | 20 | Han et al., 2007 [46] |
| Tsf | Elongation factor Ts | 30.6 | Han et al., 2007 [47] |
| SlyD | Aggregation-resistant protein | 22.2 | Han et al., 2007 [48] |
| msyB | Acidic protein | 14 | Su et al., 2007 [49] |
| RpoS | RNA polymerase sigma factor | 39 | Park et al., 2008 [50] |
| yjgD | 15 | Zou et al., 2008 [51] | |
| rpoD | σ 70 factor of RNA polymerase | 20 | Zou et al., 2008 [51] |
| HaloTag7 | Inactive derivative of DhaA, a bacterial haloalkane dehalogenase | 34 | Ohana et al., 2009 [52] |
| sfGFP | Superfolder green fluorescent protein | Wu et al., 2009 [53] | |
| Mocr | Monomeric bacteriophage T7 0.3 | 16.7 | DelProposto et al., 2009 [54] |
| SNUT | Solubility eNhancing Ubiquitous Tag | 19 | Caswell et al., 2010 [55] |
| EspA | E. coli secreted protein A | 25 | Cheng et al., 2010 [56] |
| ArsC | Arsenate reductase | 16 | Song et al., 2011 [57] |
| BLA | AmpC-type β-lactamase | Tokunaga et al., 2010 [58] | |
| InfB1-21 | Entity of InfB(1-471) responsible for increased expression | 28 | Hansted et al., 2011 [59] |
| Fh8 | Fasciola hepatica antigen | 8 | Costa et al., 2014 [28] |
| SmbP | Small metal-binding protein | 9.9 | Vargas-Cortez et al., 2016 [60] |
| Ffu | β-fructofuranosidase truncations | 17.7–29.5 | Cheng et al., 2017 [61] |
| TDX | Tetracopeptide domain-containing thioredoxin | 35 | Xiao et al., 2018 [62] |
| HE-MBP(Pyr) | Truncated maltotriose-binding protein with modified histidine tag | Han et al., 2018 [63] |
| Name | pI | Size (kDa) | Reference |
|---|---|---|---|
| Polycationic tags | |||
| (Arg)1–5 | 10.00–12.62 | 0.174–0.799 | Kato et al., 2007 [67] |
| (Arg)5 (His)5 | 12.62 7.66 | 0.799 0.704 | Islam et al., 2015 [73] Islam et al., 2012 [101] Khan et al., 2015 [102] |
| (Arg)10 | 12.95 | 1.6 | Jung H-J et al., 2011 [25] Johnson et al., 2007 [97] |
| (Arg-Gly-Gly)3-Gly | 12.30 | 0.886 | Englebretsen et al., 1999 [92] |
| Poly(Arg) | Smith et al., 1984 [77] | ||
| (Gly-Arg)3-(Arg)3 | 12.91 | 1.5 | Kalpana et al., 2018 [78] |
| (Gly-Arg)4 | 12.48 | 0.871 | Englebretsen et al., 1996 [91] |
| Gly-(Arg)5 | 12.62 | 0.856 | Sato et al., 2005 [96] |
| Gly(Arg-Gly-Gly)3 Gly(Lys-Gly)6 | 12.30 10.70 | 0.886 1.2 | Choma et al., 1998 [93] |
| (Gly)2-(Arg)2-Gly-Arg Gly-Lys-Gly-(Lys)2 | 12.30 10.28 | 0.658 0.517 | Gao et al., 2017 |
| (Gly)2(Lys)4 | 10.47 | 0.645 | Park et al., 2003 [76] |
| (Lys)1–5 | 8.88–10.61 | 0.146–0.659 | Kato et al., 2007 [67] |
| (Lys)2 | 10.00 | 0.274 | Wang et al., 1999 [75] |
| (Lys)3 | 10.28 | 0.402 | Bianchi et al., 1994 [71] |
| (Lys)5 | 10.61 | 0.659 | Islam et al., 2015 [73] Hossain et al., 2009 [94] Islam et al., 2012 [101] Khan et al., 2015 [102] |
| (Lys)6 | 10.70 | 0.787 | Hage et al., 2015 [27] |
| (Lys)10 | 10.94 | 1.3 | Englebretsen et al., 1999 [92] |
| Polyanionic tags | |||
| (Asp)5 | 3.34 | 0.593 | Kim et al., 2015 [99] Kim et al., 2014 [104] |
| (Asp)5 (Glu)5 | 3.34 | 0.664 | Islam et al., 2015 [73] Islam et al., 2012 [101] Khan et al., 2015 [102] |
| [Gly-(Asp)3]3 | 3.16 | 1.2 | Rathnayaka et al., 2011 [98] |
| Negative peptide extensions (>−6) | Zhang et al., 2004 [100] | ||
| Polar tags | |||
| (Asn)5 (Gln)5 (Ser)5 | 5.50 5.50 5.50 | 0.588 0.659 0.453 | Islam et al., 2015 [73] Islam et al., 2012 [101] Khan et al., 2015 [102] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paraskevopoulou, V.; Falcone, F.H. Polyionic Tags as Enhancers of Protein Solubility in Recombinant Protein Expression. Microorganisms 2018, 6, 47. https://doi.org/10.3390/microorganisms6020047
Paraskevopoulou V, Falcone FH. Polyionic Tags as Enhancers of Protein Solubility in Recombinant Protein Expression. Microorganisms. 2018; 6(2):47. https://doi.org/10.3390/microorganisms6020047
Chicago/Turabian StyleParaskevopoulou, Vasiliki, and Franco H. Falcone. 2018. "Polyionic Tags as Enhancers of Protein Solubility in Recombinant Protein Expression" Microorganisms 6, no. 2: 47. https://doi.org/10.3390/microorganisms6020047
APA StyleParaskevopoulou, V., & Falcone, F. H. (2018). Polyionic Tags as Enhancers of Protein Solubility in Recombinant Protein Expression. Microorganisms, 6(2), 47. https://doi.org/10.3390/microorganisms6020047





