Comparing the Recombinant Protein Production Potential of Planktonic and Biofilm Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Flow Cell System and Experimental Conditions
2.3. Biofilm and Planktonic Monitoring
2.4. Measurement of Plasmid Maintenance
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Overton, T.W. Recombinant protein production in bacterial hosts. Drug Discov. Today 2014, 19, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L.; Vaishnav, P. Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 2009, 27, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Gopal, G.J.; Kumar, A. Strategies for the production of recombinant protein in Escherichia coli. Protein J. 2013, 32, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Johnson, I.S. Human insulin from recombinant DNA technology. Science 1983, 219, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Garcia, L.; Martín, L.; Mangues, R.; Ferrer-Miralles, N.; Vázquez, E.; Villaverde, A. Recombinant pharmaceuticals from microbial cells: A 2015 update. Microb. Cell Fact. 2016, 15, 33. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881. [Google Scholar] [CrossRef] [PubMed]
- Cook, L.C.; Dunny, G.M. The influence of biofilms in the biology of plasmids. Microbiol. Spectrum 2014, 2, 12. [Google Scholar]
- Beloin, C.; Ghigo, J.-M. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 2005, 13, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Mergulhão, F.J.; Monteiro, G.A.; Cabral, J.M.; Taipa, M.A. Design of bacterial vector systems for the production of recombinant proteins in Escherichia coli. J. Microbiol. Biotechnol. 2004, 14, 1–14. [Google Scholar]
- Hoffmann, F.; Rinas, U. Stress induced by recombinant protein production in Escherichia coli. In Physiological Stress Responses in Bioprocesses; Springer: Berlin, Germany, 2004; pp. 73–92. [Google Scholar]
- Andersson, L.; Yang, S.; Neubauer, P.; Enfors, S.-O. Impact of plasmid presence and induction on cellular responses in fed batch cultures of Escherichia coli. J. Biotechnol. 1996, 46, 255–263. [Google Scholar] [CrossRef]
- Ghigo, J.-M. Natural conjugative plasmids induce bacterial biofilm development. Nature 2001, 412, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Reisner, A.; Höller, B.M.; Molin, S.; Zechner, E.L. Synergistic effects in mixed Escherichia coli biofilms: Conjugative plasmid transfer drives biofilm expansion. J. Bacteriol. 2006, 188, 3582–3588. [Google Scholar] [CrossRef] [PubMed]
- May, T.; Okabe, S.J. Escherichia coli harboring a natural IncF conjugative F plasmid develops complex mature biofilms by stimulating synthesis of colanic acid and curli. J. Bacteriol. 2008, 190, 7479–7490. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ma, Q.; Wood, T.K. The R1 conjugative plasmid increases Escherichia coli biofilm formation through an envelope stress response. Appl. Environ. Microbiol. 2008, 74, 2690–2699. [Google Scholar] [CrossRef] [PubMed]
- Burmølle, M.; Bahl, M.I.; Jensen, L.B.; Sørensen, S.J.; Hansen, L.H. Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology 2008, 154, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Teodósio, J.; Simões, M.; Mergulhão, F. The influence of nonconjugative Escherichia coli plasmids on biofilm formation and resistance. J. Appl. Microbiol. 2012, 113, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.; La, H.J.; Sheng, H.; Forney, L.J.; Hovde, C.J. Influence of plasmid pO157 on Escherichia coli O157:H7 Sakai biofilm formation. Appl. Environ. Microbiol. 2010, 76, 963–966. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.T.; Peretti, S.W.; Bryers, J.D. Plasmid retention and gene expression in suspended and biofilm cultures of recombinant Escherichia coli DH5α (pMJR1750). Biotechnol. Bioeng. 1993, 41, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.T.; Peretti, S.W.; Bryers, J.D. Effects of medium carbon-to-nitrogen ratio on biofilm formation and plasmid stability. Biotechnol. Bioeng. 1994, 44, 329–336. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, H.A.; Niu, C.; Gilbert, E.S. Enhanced high copy number plasmid maintenance and heterologous protein production in an Escherichia coli biofilm. Biotechnol. Bioeng. 2007, 97, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Gomes, L.; Mergulhão, F. Heterologous protein production in Escherichia coli biofilms: A non-conventional form of high cell density cultivation. Process Biochem. 2017, 57, 1–8. [Google Scholar] [CrossRef]
- Mergulhão, F.J.; Monteiro, G.A. Analysis of factors affecting the periplasmic production of recombinant proteins in Escherichia coli. J. Microbiol. Biotechnol. 2007, 17, 1236. [Google Scholar] [PubMed]
- Gomes, L.; Carvalho, D.; Briandet, R.; Mergulhão, F. Temporal variation of recombinant protein expression in Escherichia coli biofilms analysed at single-cell level. Process Biochem. 2016, 51, 1155–1161. [Google Scholar] [CrossRef]
- Teodósio, J.; Simões, M.; Melo, L.; Mergulhão, F. Flow cell hydrodynamics and their effects on E. coli biofilm formation under different nutrient conditions and turbulent flow. Biofouling 2011, 27, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.-X.; Qian, Z.-G.; Ki, C.S.; Park, Y.H.; Kaplan, D.L.; Lee, S.Y. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc. Natl. Acad. Sci. USA 2010, 107, 14059–14063. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-X.; Qian, Z.-G.; Zhong, J.-J.; Xia, X.-X. Hyper-production of large proteins of spider dragline silk MaSp2 by Escherichia coli via synthetic biology approach. Process Biochem. 2016, 51, 484–490. [Google Scholar] [CrossRef]
- Diaz Ricci, J.C.; Hernández, M.E. Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 2000, 20, 79–108. [Google Scholar] [CrossRef] [PubMed]
- Kurland, C.; Dong, H. Bacterial growth inhibition by overproduction of protein. Mol. Microbiol. 1996, 21, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-T.; Peretti, S.W.; Bryers, J.D. Effects of inducer levels on a recombinant bacterial biofilm formation and gene expression. Biotechnol. Lett. 1994, 16, 903–908. [Google Scholar] [CrossRef]
- Williams, I.; Venables, W.A.; Lloyd, D.; Paul, F.; Critchley, I. The effects of adherence to silicone surfaces on antibiotic susceptibility in Staphylococcus aureus. Microbiology 1997, 143, 2407–2413. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.G.; Geesey, G.G. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl. Environ. Microbiol. 1995, 61, 860–867. [Google Scholar] [PubMed]
- May, T.; Ito, A.; Okabe, S. Induction of multidrug resistance mechanism in Escherichia coli biofilms by interplay between tetracycline and ampicillin resistance genes. Antimicrob. Agents Chemother. 2009, 53, 4628–4639. [Google Scholar] [CrossRef] [PubMed]
- Cook, L.C.; Dunny, G.M. Effects of biofilm growth on plasmid copy number and expression of antibiotic resistance genes in Enterococcus faecalis. Antimicrob. Agents Chemother. 2013, 57, 1850–1856. [Google Scholar] [CrossRef] [PubMed]
- Bongaerts, R.J.; Hautefort, I.; Sidebotham, J.M.; Hinton, J.C. Green fluorescent protein as a marker for conditional gene expression in bacterial cells. In Methods in Enzymology; Clark, V.L., Patrik, M.B., Eds.; Elsevier: New York, NY, USA, 2002; Volume 358, pp. 43–66. [Google Scholar]
- Naresh, K. Applications of fluorescence spectroscopy. J. Chem. Pharm. Sci. 2014, 974, 2115. [Google Scholar]
- Webb, D.J.; Brown, C.M. Epi-fluorescence microscopy. In Cell Imaging Techniques; Taatjes, D., Roth, J., Eds.; Springer; Humana Press: Totowa, NJ, USA, 2012; pp. 29–59. [Google Scholar]
 ) and squares (
) and squares (  ) on the right y-axis. The means ± SDs for three independent experiments are illustrated.
) on the right y-axis. The means ± SDs for three independent experiments are illustrated.
 ) and squares (
) and squares (  ) on the right y-axis. The means ± SDs for three independent experiments are illustrated.
) on the right y-axis. The means ± SDs for three independent experiments are illustrated.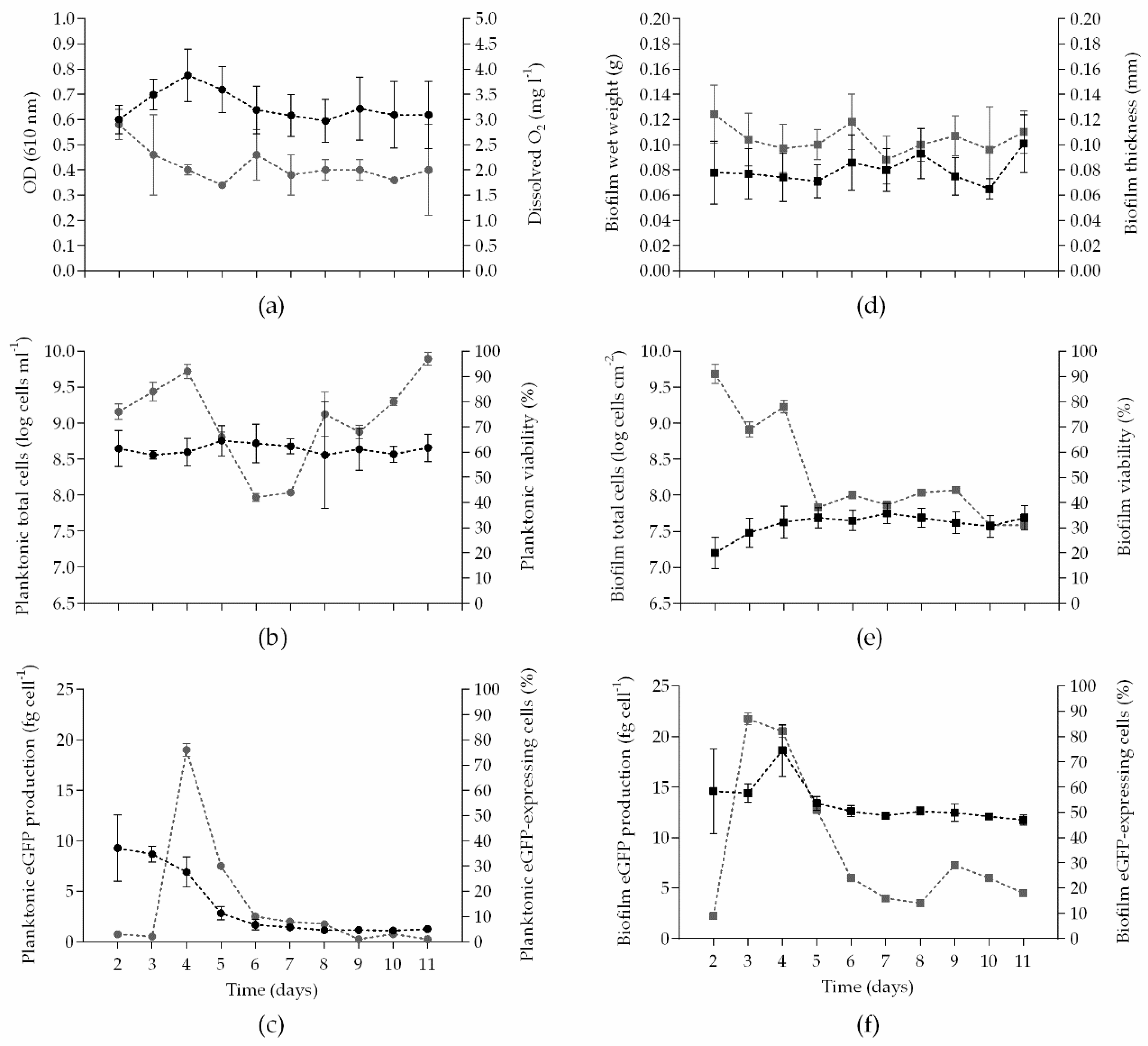
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares, A.; Gomes, L.C.; Mergulhão, F.J. Comparing the Recombinant Protein Production Potential of Planktonic and Biofilm Cells. Microorganisms 2018, 6, 48. https://doi.org/10.3390/microorganisms6020048
Soares A, Gomes LC, Mergulhão FJ. Comparing the Recombinant Protein Production Potential of Planktonic and Biofilm Cells. Microorganisms. 2018; 6(2):48. https://doi.org/10.3390/microorganisms6020048
Chicago/Turabian StyleSoares, Alexandra, Luciana Calheiros Gomes, and Filipe José Mergulhão. 2018. "Comparing the Recombinant Protein Production Potential of Planktonic and Biofilm Cells" Microorganisms 6, no. 2: 48. https://doi.org/10.3390/microorganisms6020048




