Growth and Cell Size of Microalga Auxenochlorella protothecoides AS-1 under Different Trophic Modes
Abstract
:1. Introduction
2. Materials and Methods
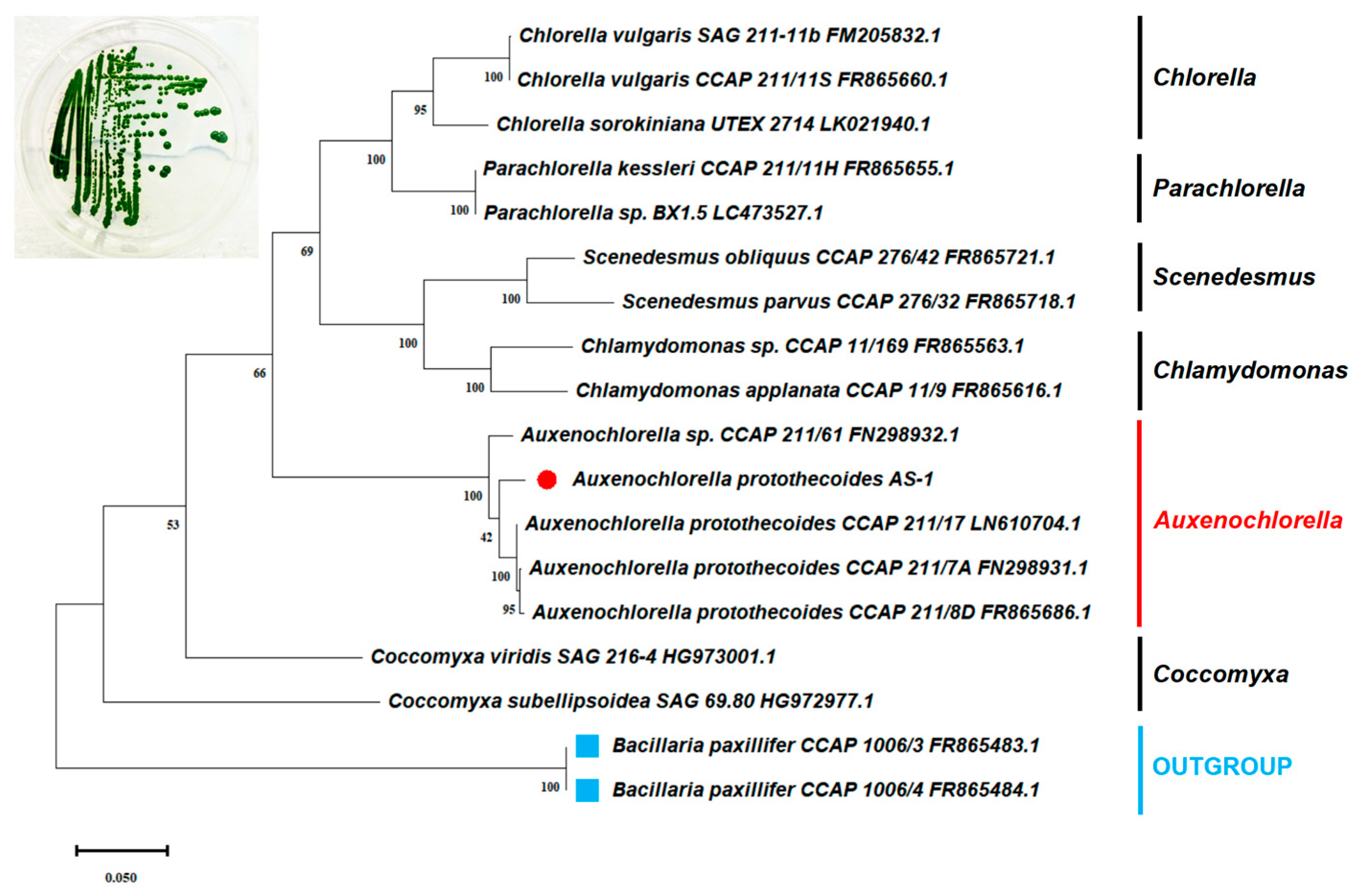
2.1. Isolation of Green Algal Strain AS-1
2.2. Identification of Strain AS-1 Based on the Sequence of the 18S rRNA-ITS1-5.8S rRNA-ITS2 Operon
2.3. Growth of A. protothecoides AS-1 under Different Trophic Modes
2.4. Growth Measurement
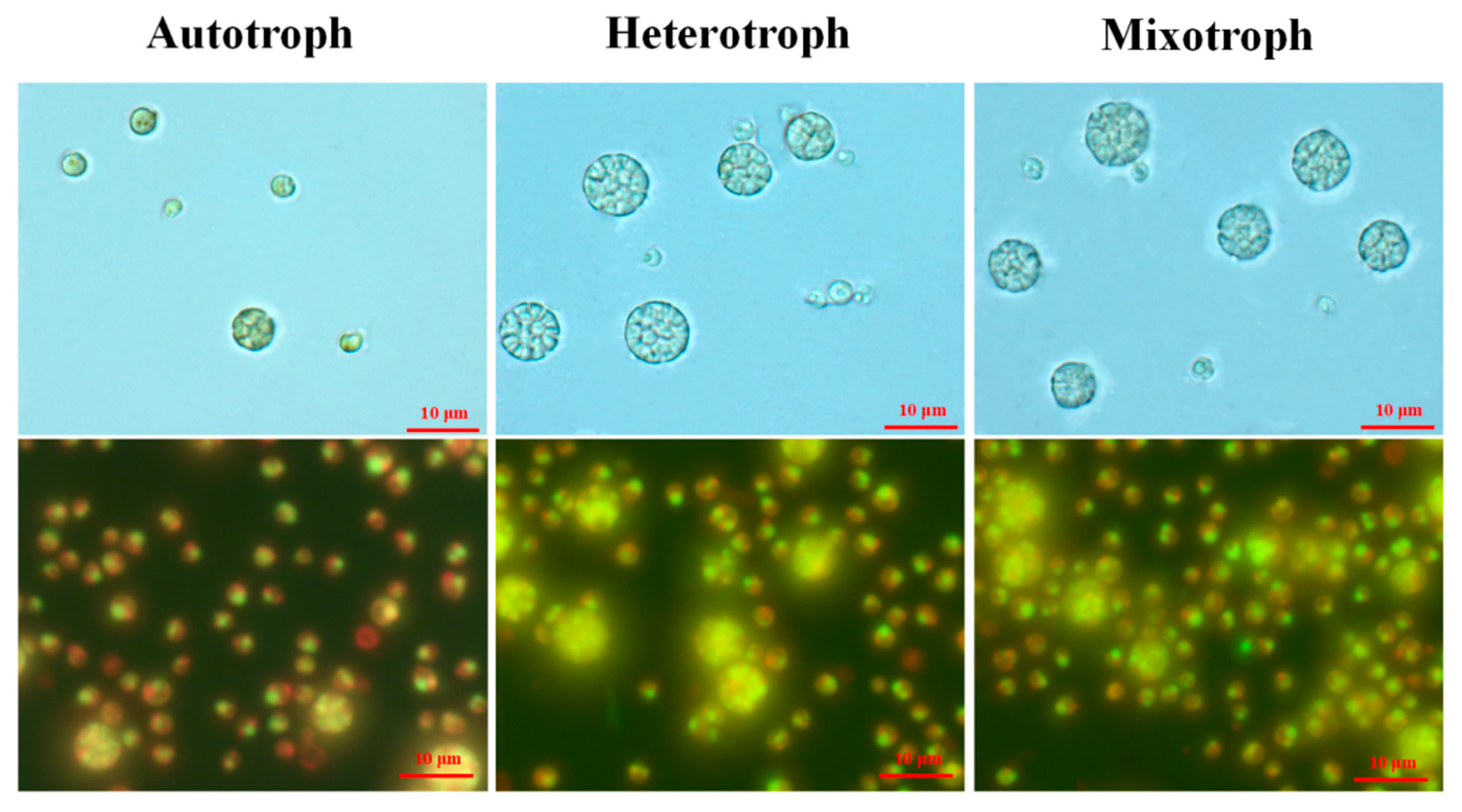
2.5. Observation of Algal Cells with Light and Fluorescence Microscope
2.6. Cell Size Measurement with the Coulter Counter
2.7. Data and Statistical Analysis
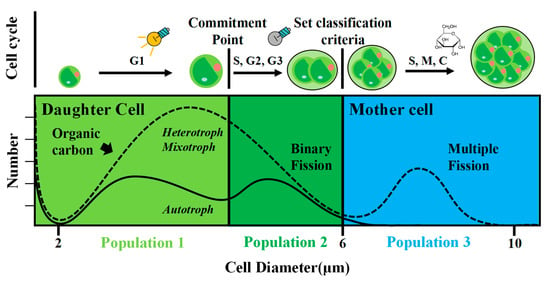
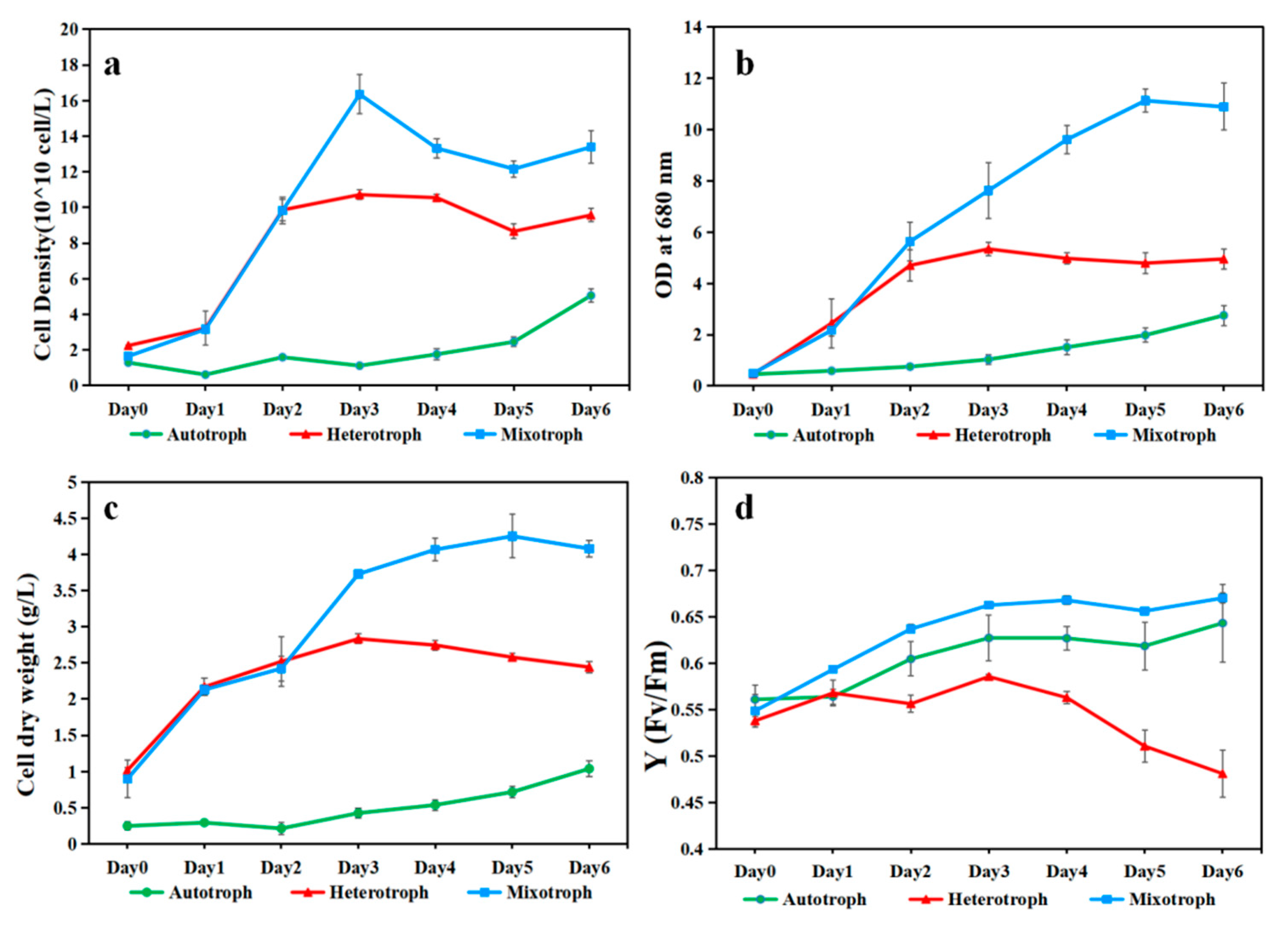
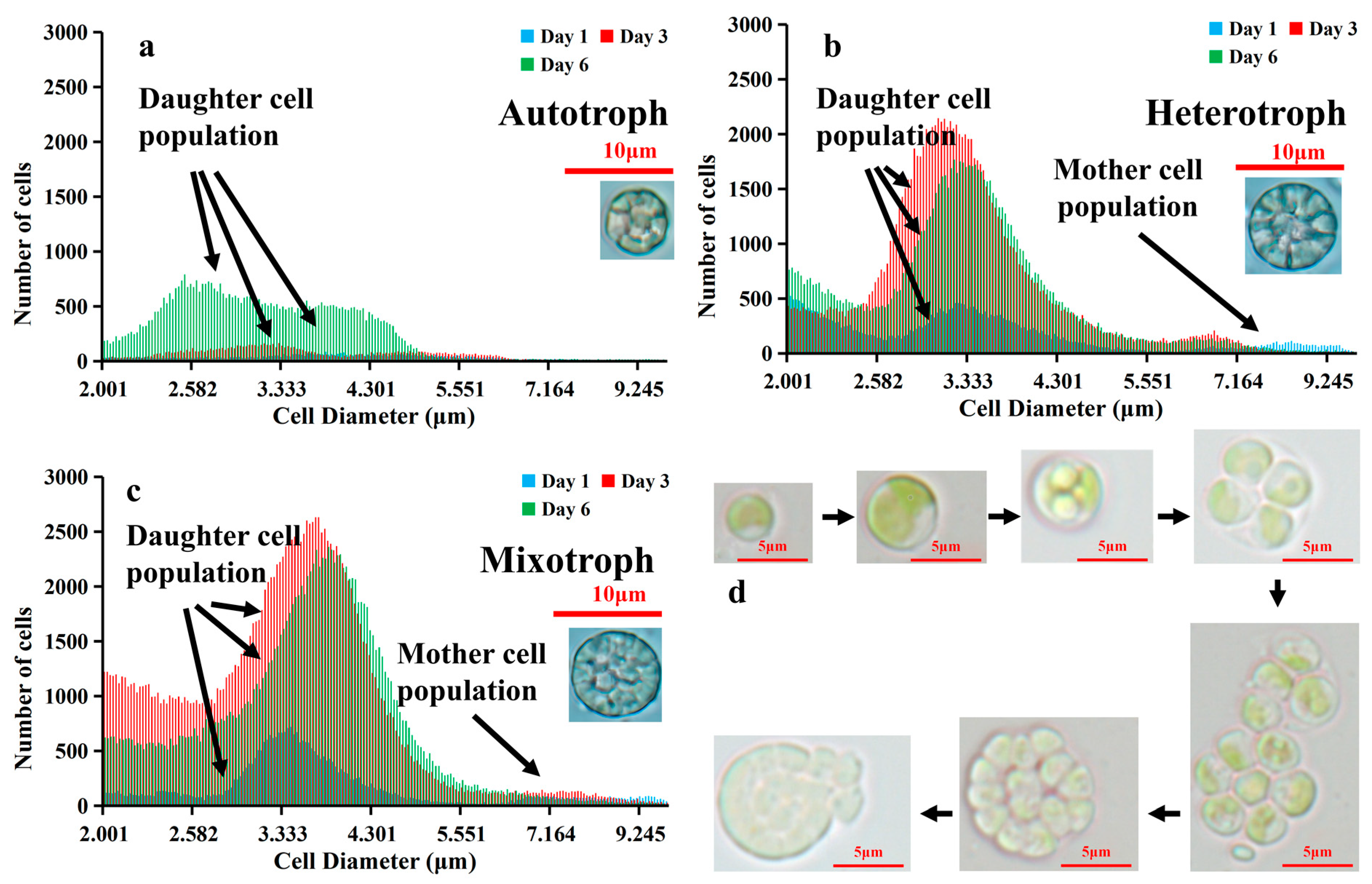
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Spolaore, P.; Joannis-Cassan, C.; Duran, E.; Isambert, A. Commercial applications of microalgae. J. Biosci. Bioeng. 2006, 101, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Del Campo, J.A.; García-González, M.; Guerrero, M.G. Outdoor cultivation of microalgae for carotenoid production: Current state and perspectives. Appl. Microbiol. Biotechnol. 2007, 74, 1163–1174. [Google Scholar] [CrossRef] [PubMed]
- Dineshbabu, G.; Goswami, G.; Kumar, R.; Sinha, A.; Das, D. Microalgae–nutritious, sustainable aqua-and animal feed source. J. Funct. Foods 2019, 62, 103545. [Google Scholar] [CrossRef]
- Singh, S.; Kate, B.N.; Banerjee, U.C. Bioactive compounds from cyanobacteria and microalgae: An overview. Crit. Rev. Biotechnol. 2005, 25, 73–95. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, J.E.; Cho, Y.B.; Hwang, S.J. Growth rate, organic carbon, and nutrient removal rates of Chlorella sorokiniana in autotrophic, heterotrophic and mixotrophic conditions. Bioresour. Technol. 2013, 144, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Morales-Sánchez, D.; Martinez-Rodriguez, O.A.; Martinez, A. Heterotrophic cultivation of microalgae: Production of metabolites of commercial interest. J. Chem. Technol. Biotechnol. 2017, 92, 925–936. [Google Scholar] [CrossRef]
- Kong, W.; Yang, S.; Wang, H.; Huo, H.; Guo, B.; Liu, N.; Niu, S. Regulation of biomass, pigments, and lipid production by Chlorella vulgaris 31 through controlling trophic modes and carbon sources. J. Appl. Phycol. 2020, 32, 1569–1579. [Google Scholar] [CrossRef]
- Droop, M.R. Heterotrophy of carbon. Algal Physiol. Biochem. 1974, 530–559. [Google Scholar]
- Nicodemou, A.; Kallis, M.; Agapiou, A.; Markidou, A.; Koutinas, M. The effect of trophic modes on biomass and lipid production of five microalgal strains. Water 2022, 14, 240. [Google Scholar] [CrossRef]
- Yamamoto, M.; Nozaki, H.; Kawano, S. Evolutionary relationships among multiple modes of cell division in the genus Nannochloris (Chlorophyta) revealed by genome size, actin gene multiplicity, and phylogeny. J. Phycol. 2001, 37, 106–120. [Google Scholar] [CrossRef]
- Bišová, K.; Zachleder, V. Cell-cycle regulation in green algae dividing by multiple fission. J. Exp. Bot. 2014, 65, 2585–2602. [Google Scholar] [CrossRef] [PubMed]
- Zachleder, V.; Bišová, K.; Vítová, M. The cell cycle of microalgae. Physiol. Microalgae 2016, 6, 3–46. [Google Scholar]
- Tamiya, H.; Iwamura, T.; Shibata, K.; Hase, E.; Nihei, T. Correlation between photosynthesis and light-independent metabolism in the growth of Chlorella. Biochim. Biophys. Acta 1953, 12, 23–40. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.W. The nuclear cell cycle in Chlamydomonas (Chlorophyceae) 1. J. Phycol. 1982, 18, 192–195. [Google Scholar] [CrossRef]
- Yamamoto, M.; Fujishita, M.; Hirata, A.; Kawano, S. Regeneration and maturation of daughter cell walls in the autospore-forming green alga Chlorella vulgaris (Chlorophyta, Trebouxiophyceae). J. Plant Res. 2004, 117, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. r-and K-tactics in the evolution of protist developmental systems: Cell and genome size, phenotype diversifying selection, and cell cycle patterns. Biosystems 1980, 12, 43–59. [Google Scholar] [CrossRef] [PubMed]
- Fenchel, T. Intrinsic rate of natural increase: The relationship with body size. Oecologia 1974, 14, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Warwick, R.M.; Joint, I.R. The size distribution of organisms in the Celtic Sea: From bacteria to Metazoa. Oecologia 1987, 73, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Gaedke, U. The size distribution of plankton biomass in a large lake and its seasonal variability. Limnol. Oceanogr. 1992, 37, 1202–1220. [Google Scholar] [CrossRef]
- Chakraborty, S.; Nielsen, L.T.; Andersen, K.H. Trophic strategies of unicellular plankton. Am. Nat. 2017, 189, E77–E90. [Google Scholar] [CrossRef]
- Ho, P.C.; Chang, C.W.; Shiah, F.K.; Wang, P.L.; Hsieh, C.H.; Andersen, K.H. Body size, light intensity, and nutrient supply determine plankton stoichiometry in mixotrophic plankton food webs. Am. Nat. 2020, 195, E100–E111. [Google Scholar] [CrossRef] [PubMed]
- Hillebrand, H.; Acevedo-Trejos, E.; Moorthi, S.D.; Ryabov, A.; Striebel, M.; Thomas, P.K.; Schneider, M.L. Cell size as driver and sentinel of phytoplankton community structure and functioning. Funct. Ecol. 2022, 36, 276–293. [Google Scholar] [CrossRef]
- Concas, A.; Pisu, M.; Cao, G. A novel mathematical model to simulate the size-structured growth of microalgae strains dividing by multiple fission. Chem. Eng. J. 2016, 287, 252–268. [Google Scholar] [CrossRef]
- Rioboo, C.; O’Connor, J.E.; Prado, R.; Herrero, C.; Cid, Á. Cell proliferation alterations in Chlorella cells under stress conditions. Aquat. Toxicol. 2009, 94, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Javanmardian, M.; Palsson, B.O. Continuous photoautotrophic cultures of the eukaryotic alga Chlorella vulgaris can exhibit stable oscillatory dynamics. Biotechnol. Bioeng. 1992, 39, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Craigie, R.A.; Cavalier-Smith, T. Cell volume and the control of the Chlamydomonas cell cycle. J. Cell Sci. 1982, 54, 173–191. [Google Scholar] [CrossRef]
- Oldenhof, H.; Zachleder, V.; Van Den Ende, H. Blue-and red-light regulation of the cell cycle in Chlamydomonas reinhardtii (Chlorophyta). Eur. J. Phycol. 2006, 41, 313–320. [Google Scholar] [CrossRef]
- Jong, L.W.; Fujiwara, T.; Hirooka, S.; Miyagishima, S.Y. Cell size for commitment to cell division and number of successive cell divisions in Cyanidialean red algae. Protoplasma 2021, 258, 1103–1118. [Google Scholar] [CrossRef]
- Loveland, P.J.; Whalley, W.R. Particle size analysis. In Soil and Environmental Analysis; CRC Press: Boca Raton, FL, USA, 2000; pp. 293–326. [Google Scholar]
- Krediet, C.J.; DeNofrio, J.C.; Caruso, C.; Burriesci, M.S.; Cella, K.; Pringle, J.R. Rapid, precise, and accurate counts of Symbiodinium cells using the guava flow cytometer, and a comparison to other methods. PLoS ONE 2015, 10, e0135725. [Google Scholar] [CrossRef]
- Sosa, A.P. Microbial Biofilms on Microplastics: A Look into the Estuarine Plastisphere of the Chesapeake Bay. Ph.D. Thesis, University of Maryland, College Park, MD, USA, 2021. [Google Scholar]
- Kan, J. Bacterioplankton in the Chesapeake Bay: Genetic Diversity, Population Dynamics, and Community Proteomics. Ph.D. Thesis, University of Maryland, College Park, MD, USA, 2006. [Google Scholar]
- Patel, A.K.; Joun, J.M.; Hong, M.E.; Sim, S.J. Effect of light conditions on Mixotrophic cultivation of green microalgae. Bioresour. Technol. 2019, 282, 245–253. [Google Scholar] [CrossRef]
- Vítová, M.; Hendrychová, J.; Cepák, V.; Zachleder, V. Visualization of DNA-containing structures in various species of Chlorophyta, Rhodophyta and Cyanophyta using SYBR Green I dye. Folia Microbiol. 2005, 50, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Guiry, M.D.; Guiry, G.M. AlgaeBase. World-Wide Electronic Publication, University of Galway. Available online: https://www.algaebase.org (accessed on 9 March 2024).
- Shihira, I.; Krauss, R.W. Chlorella, Physiology, and Taxonomy of Forty-One Isolates; NASA-CR-69107; NASA: Washington, DC, USA, 1965. [Google Scholar]
- Kalina, T.; Puncochárová, M. Taxonomy of the subfamily Scotiellocystoideae Fott 1976 (Chlorellaceae, Chlorophyceae). Arch. Hydrobiol. Suppl. Algol. Stud. 1987, 45, 473–521. [Google Scholar]
- Yan, D.; Wang, Y.; Murakami, T.; Shen, Y.; Gong, J.; Jiang, H.; Wu, Q. Auxenochlorella protothecoides and Prototheca wickerhamii plastid genome sequences give insight into the origins of non-photosynthetic algae. Sci. Rep. 2015, 5, 14465. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Jian, J.; Wang, L.; Xiong, L.; Lin, H.; Zhou, Z.; Wu, W. Genome sequences of two strains of Prototheca wickerhamii provide insight into the Protothecosis evolution. Front. Cell. Infect. Microbiol. 2022, 12, 797017. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Wu, Q. Biodiesel production from heterotrophic microalgal oil. Bioresour. Technol. 2006, 97, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Miao, X.; Wu, Q. High quality biodiesel production from a microalga Chlorella protothecoides by heterotrophic growth in fermenters. J. Biotechnol. 2006, 126, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Krzemińska, I.; Oleszek, M. Glucose supplementation-induced changes in the Auxenochlorella protothecoides fatty acid composition suitable for biodiesel production. Bioresour. Technol. 2016, 218, 1294–1297. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Min, M.; Zhou, W.; Du, Z.; Mohr, M.; Chen, P.; Ruan, R. Enhanced mixotrophic growth of microalga Chlorella sp. on pretreated swine manure for simultaneous biofuel feedstock production and nutrient removal. Bioresour. Technol. 2012, 126, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Li, Y.; Min, M.; Hu, B.; Zhang, H.; Ma, X.; Ruan, R. Growing wastewater-born microalga Auxenochlorella protothecoides UMN280 on concentrated municipal wastewater for simultaneous nutrient removal and energy feedstock production. Appl. Energy 2012, 98, 433–440. [Google Scholar] [CrossRef]
- Ramos Tercero, E.A.; Sforza, E.; Morandini, M.; Bertucco, A. Cultivation of Chlorella protothecoides with urban wastewater in continuous photobioreactor: Biomass productivity and nutrient removal. Appl. Biochem. Biotechnol. 2014, 172, 1470–1485. [Google Scholar] [CrossRef]
- Abreu, A.P.; Fernandes, B.; Vicente, A.A.; Teixeira, J.; Dragone, G. Mixotrophic cultivation of Chlorella vulgaris using industrial dairy waste as organic carbon source. Bioresour. Technol. 2012, 118, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Cheirsilp, B.; Torpee, S. Enhanced growth and lipid production of microalgae under mixotrophic culture condition: Effect of light intensity, glucose concentration and fed-batch cultivation. Bioresour. Technol. 2012, 110, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Girard, J.M.; Roy, M.L.; Hafsa, M.B.; Gagnon, J.; Faucheux, N.; Heitz, M.; Deschênes, J.S. Mixotrophic cultivation of green microalgae Scenedesmus obliquus on cheese whey permeate for biodiesel production. Algal Res. 2014, 5, 241–248. [Google Scholar] [CrossRef]
- Li, Y.R.; Tsai, W.T.; Hsu, Y.C.; Xie, M.Z.; Chen, J.J. Comparison of autotrophic and mixotrophic cultivation of green microalgal for biodiesel production. Energy Procedia 2014, 52, 371–376. [Google Scholar] [CrossRef]
- Yu, Q.; Wang, H.; Li, X.; Yin, Y.; Qin, S.; Ge, B. Enhanced biomass and CO2 sequestration of Chlorella vulgaris using a new mixotrophic cultivation method. Process Biochem. 2020, 90, 168–176. [Google Scholar] [CrossRef]
- Heredia-Arroyo, T.; Wei, W.; Hu, B. Oil accumulation via heterotrophic/mixotrophic Chlorella protothecoides. Appl. Biochem. Biotechnol. 2010, 162, 1978–1995. [Google Scholar] [CrossRef] [PubMed]
- Caporgno, M.P.; Haberkorn, I.; Böcker, L.; Mathys, A. Cultivation of Chlorella protothecoides under different growth modes and its utilization in oil/water emulsions. Bioresour. Technol. 2019, 288, 121476. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Wang, Y.; Shen, Y.; Yan, D.; He, X.; Dai, J.; Wu, Q. Oil accumulation mechanisms of the oleaginous microalga Chlorella protothecoides revealed through its genome, transcriptomes, and proteomes. BMC Genom. 2014, 15, 1–14. [Google Scholar] [CrossRef]
- Perez-Garcia, O.; Escalante, F.M.; De-Bashan, L.E.; Bashan, Y. Heterotrophic cultures of microalgae: Metabolism and potential products. Water Res. 2011, 45, 11–36. [Google Scholar] [CrossRef]
- Wu, Z.; Shi, X. Optimization for high density cultivation of heterotrophic Chlorella based on a hybrid neural network model. Lett. Appl. Microbiol. 2007, 44, 13–18. [Google Scholar] [CrossRef]
- Cross, F.R.; Umen, J.G. The Chlamydomonas cell cycle. Plant J. 2015, 82, 370–392. [Google Scholar] [CrossRef] [PubMed]
- Tamiya, H. Synchronous cultures of algae. Annu. Rev. Plant Physiol. 1966, 17, 1–27. [Google Scholar] [CrossRef]
- Zachleder, V.; Ende, H.V.D. Cell cycle events in the green alga Chlamydomonas eugametos and their control by environmental factors. J. Cell Sci. 1992, 102, 469–474. [Google Scholar] [CrossRef]
- He, X.; Dai, J.; Wu, Q. Identification of sporopollenin as the outer layer of cell wall in microalga Chlorella protothecoides. Front. Microbiol. 2016, 7, 198163. [Google Scholar] [CrossRef] [PubMed]
- Rading, M.M.; Engel, T.A.; Lipowsky, R.; Valleriani, A. Stationary size distributions of growing cells with binary and multiple cell division. J. Stat. Phys. 2011, 145, 1–22. [Google Scholar] [CrossRef]
- Zhao, L.; Dai, J.; Wu, Q. Autophagy-like processes are involved in lipid droplet degradation in Auxenochlorella protothecoides during the heterotrophy-autotrophy transition. Front. Plant Sci. 2014, 5, 400. [Google Scholar] [CrossRef] [PubMed]
- Chioccioli, M.; Hankamer, B.; Ross, I.L. Flow cytometry pulse width data enables rapid and sensitive estimation of biomass dry weight in the microalgae Chlamydomonas reinhardtii and Chlorella vulgaris. PLoS ONE 2014, 9, e97269. [Google Scholar] [CrossRef]
- Sánchez-Alvarez, E.L.; González-Ledezma, G.; Prats, J.A.B.; Stephano-Hornedo, J.L.; Hildebrand, M. Evaluating Marinichlorella kaistiae KAS603 cell size variation, growth and TAG accumulation resulting from rapid adaptation to highly diverse trophic and salinity cultivation regimes. Algal Res. 2017, 25, 12–24. [Google Scholar] [CrossRef]
- Li, T.; Yang, F.; Xu, J.; Wu, H.; Mo, J.; Dai, L.; Xiang, W. Evaluating differences in growth, photosynthetic efficiency, and transcriptome of Asterarcys sp. SCS-1881 under autotrophic, mixotrophic, and heterotrophic culturing conditions. Algal Res. 2020, 45, 101753. [Google Scholar] [CrossRef]
- Marañón, E.; Cermeño, P.; López-Sandoval, D.C.; Rodríguez-Ramos, T.; Sobrino, C.; Huete-Ortega, M.; Rodríguez, J. Unimodal size scaling of phytoplankton growth and the size dependence of nutrient uptake and use. Ecol. Lett. 2013, 16, 371–379. [Google Scholar] [CrossRef]
- Stolte, W.; Riegman, R. Effect of phytoplankton cell size on transient-state nitrate and ammonium uptake kinetics. Microbiology 1995, 141, 1221–1229. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Sosa, A.; Chen, F. Growth and Cell Size of Microalga Auxenochlorella protothecoides AS-1 under Different Trophic Modes. Microorganisms 2024, 12, 835. https://doi.org/10.3390/microorganisms12040835
Chen H, Sosa A, Chen F. Growth and Cell Size of Microalga Auxenochlorella protothecoides AS-1 under Different Trophic Modes. Microorganisms. 2024; 12(4):835. https://doi.org/10.3390/microorganisms12040835
Chicago/Turabian StyleChen, Haoyu, Ana Sosa, and Feng Chen. 2024. "Growth and Cell Size of Microalga Auxenochlorella protothecoides AS-1 under Different Trophic Modes" Microorganisms 12, no. 4: 835. https://doi.org/10.3390/microorganisms12040835