Hospital Drains as Reservoirs of Pseudomonas aeruginosa: Multiple-Locus Variable-Number of Tandem Repeats Analysis Genotypes Recovered from Faucets, Sink Surfaces and Patients
Abstract
:1. Introduction
2. Results
2.1. Culture Protocol Comparison for Environmental Samples
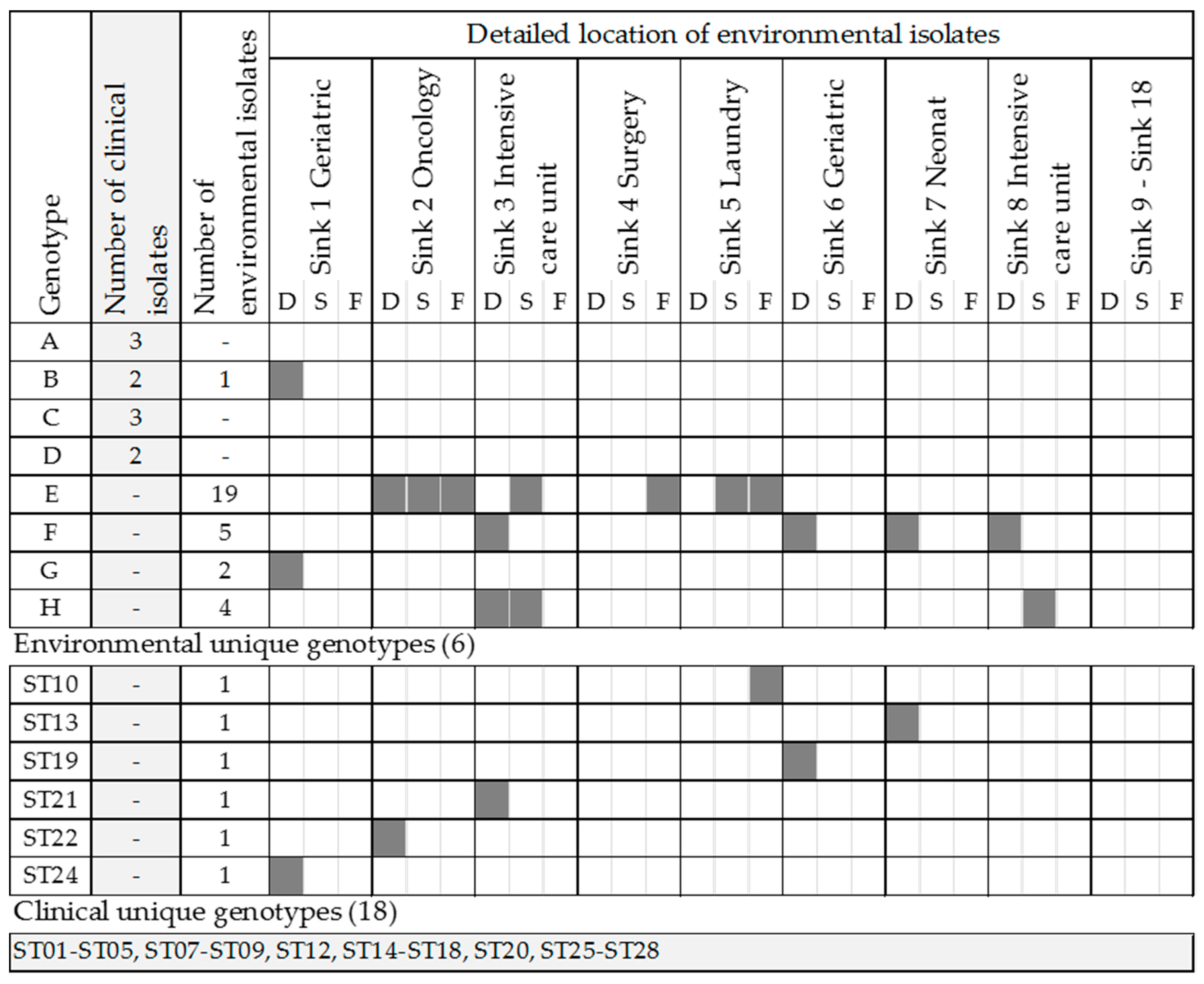
2.2. Genotyping of Environmental and Clinical Strains
2.3. Occurrence and Relationship between Sink Partsthis is an Example of an Equation
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Heatlh Organization (WHO). Report on the Burden of Endemic Health Care-Associated Infection Worldwide: A Systematic Review of the Litterature; World Heatlh Organization: Geneva, Switzerland, 2011. [Google Scholar]
- Hidron, A.I.; Edwards, J.R.; Patel, J.; Horan, T.C.; Sievert, D.M.; Pollock, D.A.; Fridkin, S.K. Nhsn annual update: Antimicrobial resistant pathogens associated with healthcare associated infections: Annual summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 2008, 29, 996–1011. [Google Scholar] [CrossRef] [PubMed]
- The RAISIN Working Group. Surveillance and outbreak reports “raisin”—A national programme for early warning, investigation and surveillance of healthcare-associated infection in France. Eurosurveillance 2009, 14, 429–433. [Google Scholar]
- Zarb, P.; Coignard, B.; Griskeviciene, J.; Muller, A.; Vankerckhoven, V.; Weist, K.; Goossens, M.M.; Vaerenberg, S.; Hopkins, S.; Catry, B.; et al. The European centre for disease prevention and control (ECDC) pilot point prevalence survey of healthcare-associated infections and antimicrobial use. Eurosurveillance 2012, 17, 4–19. [Google Scholar]
- Ferroni, A.; Nguyen, L.; Pron, B.; Quesne, G.; Brusset, M.-C.; Berche, P. Outbreak of nosocomial urinary tract infections due to Pseudomonas aeruginosa in a paediatric surgical unit associated with tap-water contamination. J. Hosp. Infect. 1998, 39, 301–307. [Google Scholar] [CrossRef]
- Yapicioglu, H.; Gokmen, T.G.; Yildizdas, D.; Koksal, F.; Ozlu, F.; Kale-Cekinmez, E.; Mert, K.; Mutlu, B.; Satar, M.; Narli, N.; et al. Pseudomonas aeruginosa infections due to electronic faucets in a neonatal intensive care unit. J. Paediatr. Child Health 2011, 48, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, D.; Terashita, D.; English, T. An Outbreak of Pseudomonas aeruginosa in Neonatal Intensive Care Unit, Los Angeles County, Special Studies Report 2006; Acute Communicable Disease Control Program: Los Angeles, CA, USA, 2006; pp. 53–55. [Google Scholar]
- Bedard, E.; Laferriere, C.; Charron, D.; Lalancette, C.; Renaud, C.; Desmarais, N.; Deziel, E.; Prevost, M. Post-outbreak investigation of Pseudomonas aeruginosa faucet contamination by quantitative polymerase chain reaction and environmental factors affecting positivity. Infect. Control Hosp. Epidemiol. 2015, 36, 1337–1343. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.T.; Jhutty, A.; Parks, S.; Willis, C.; Copley, V.; Turton, J.F.; Hoffman, P.N.; Bennett, A.M. Investigation of healthcare-acquired infections associated with Pseudomonas aeruginosa biofilms in taps in neonatal units in northern ireland. J. Hosp. Infect. 2014, 86, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Wingender, J. Hygienically relevant microorganisms in biofilms of man-made water systems. In Biofilm Highlights; Flemming, H.-C., Wingender, J., Szewzyk, U., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 5, pp. 189–238. [Google Scholar]
- Trautmann, M.; Michalsky, T.; Wiedeck, H.; Radosavljevic, V.; Ruhnke, M. Tap water colonization with Pseudomonas aeruginosa in a surgical intensive care unit (ICU) and relation to Pseudomonas infections of ICU patients. Infect. Control Hosp. Epidemiol. 2001, 22, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Lavenir, R.; Sanroma, M.; Gibert, S.; Crouzet, O.; Laurent, F.; Kravtsoff, J.; Mazoyer, M.A.; Cournoyer, B. Spatio-temporal analysis of infra-specific genetic variations among a Pseudomonas aeruginosa water network hospital population: Invasion and selection of clonal complexes. J. Appl. Microbiol. 2008, 105, 1491–1501. [Google Scholar] [CrossRef] [PubMed]
- Van der Mee-Marquet, N.; Bloc, D.; Briand, L.; Besnier, J.M.; Quentin, R. Non-touch fittings in hospitals: A procedure to eradicate Pseudomonas aeruginosa contamination. J. Hosp. Infect. 2005, 60, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.H.; Olson, B.; Nathan, C.; Kabins, S.A.; Weinstein, R.A. Pseudomonas in the sinks in an intensive care unit: Relation to patients. J. Clin. Pathol. 1984, 37, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Döring, G.; Ulrich, M.; Müller, W.; Bitzer, J.; Schmidt-Koenig, L.; Münst, L.; Grupp, H.; Wolz, C.; Stern, M.; Botzenhart, K. Generation of Pseudomonas aeruginosa aerosols during handwashing from contaminated sink drains, transmission to hands of hospital personnel, and its prevention by use of a new heating device. Int. J. Hyg. Environ. Med. 1991, 191, 494–505. [Google Scholar]
- Cholley, P.; Thouverez, M.; Floret, N.; Bertrand, X.; Talon, D. The role of water fittings in intensive care rooms as reservoirs for the colonization of patients with Pseudomonas aeruginosa. Intensive Care Med. 2008, 34, 1428–1433. [Google Scholar] [CrossRef] [PubMed]
- Cuttelod, M.; Senn, L.; Terletskiy, V.; Nahimana, I.; Petignat, C.; Eggimann, P.; Bille, J.; Prod’hom, G.; Zanetti, G.; Blanc, D.S. Molecular epidemiology of Pseudomonas aeruginosa in intensive care units over a 10-year period (1998–2007). Clin. Microbiol. Infect. 2011, 17, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Leprat, R.; Denizot, V.; Bertr, X.; Talon, D. Non-touch fittings in hospitals: A possible source of Pseudomonas aeruginosa and Legionella spp. J. Hosp. Infect. 2003, 53, 77. [Google Scholar] [CrossRef] [PubMed]
- Rogues, A.M.; Boulestreau, H.; Lashéras, A.; Boyer, A.; Gruson, D.; Merle, C.; Castaing, Y.; Bébear, C.M.; Gachie, J.P. Contribution of tap water to patient colonisation with Pseudomonas aeruginosa in a medical intensive care unit. J. Hosp. Infect. 2007, 67, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Chaidez, C.; Gerba, C.P. Comparison of the microbiologic quality of point-of-use (pou)-treated water and tap water. Int. J. Environ. Health Res. 2004, 14, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Motoshima, M.; Yanagihara, K.; Fukushima, K.; Matsuda, J.; Sugahara, K.; Hirakata, Y.; Yamada, Y.; Kohno, S.; Kamihira, S. Rapid and accurate detection of Pseudomonas aeruginosa by real-time polymerase chain reaction with melting curve analysis targeting gyrb gene. Diagn. Microbiol. Infect. Dis. 2007, 58, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Vianelli, N.; Giannini, M.B.; Quarti, C.; Bucci Sabattini, M.A.; Fiacchini, M.; de Vivo, A.; Graldi, P.; Galli, S.; Nanetti, A.; Baccarani, M.; et al. Resolution of a Pseudomonas aeruginosa outbreak in a hematology unit with the use of disposable sterile water filters. Haematologica 2006, 91, 983–985. [Google Scholar] [PubMed]
- Warburton, D.W.; Bowen, B.; Konkle, A. The survival and recovery of Pseudomonas aeruginosa and its effect upon salmonellae in water: Methodology to test bottled water in canada. Can. J. Microbiol. 1994, 40, 987–992. [Google Scholar] [CrossRef] [PubMed]
- Ortolano, G.A.; McAlister, M.B.; Angelbeck, J.A.; Schaffer, J.; Russell, R.L.; Maynard, E.; Wenz, B. Hospital water point-of-use filtration: A complementary strategy to reduce the risk of nosocomial infection. Am. J. Infect. Control 2005, 33, S1–19. [Google Scholar] [CrossRef] [PubMed]
- Charron, D.; Bédard, E.; Lalancette, C.; Laferrière, C.; Prévost, M. Impact of electronic faucets and water quality on the occurrence of Pseudomonas aeruginosa in water: A multi-hospital study. Infect. Control Hosp. Epidemiol. 2015, 36, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Hota, S.; Hirji, Z.; Stockton, K.; Lemieux, C.; Dedier, H.; Wolfaardt, G.; Gardam, M.A. Outbreak of multidrug-resistant Pseudomonas aeruginosa colonization and infection secondary to imperfect intensive care unit room design. Infect. Control Hosp. Epidemiol. 2009, 30, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Johansson, E.; Welinder-Olsson, C.; Gilljam, M. Genotyping of Pseudomonas aeruginosa isolates from lung transplant recipients and aquatic environment-detected in-hospital transmission. APMIS 2014, 122, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Schneider, H.; Geginat, G.; Hogardt, M.; Kramer, A.; Durken, M.; Schroten, H.; Tenenbaum, T. Pseudomonas aeruginosa outbreak in a pediatric oncology care unit caused by an errant water jet into contaminated siphons. Pediatr. Infect. Dis. J. 2012, 31, 648–650. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.J.; Slade, O.J.; Van Hal, S.; Espedido, B.; Gordon, A.; Farhat, R.; Chan, R. Whole genome sequencing in real-time investigation and management of a Pseudomonas aeruginosa outbreak on a neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 2015, 36, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Quick, J.; Cumley, N.; Wearn, C.M.; Niebel, M.; Constantinidou, C.; Thomas, C.M.; Pallen, M.J.; Moiemen, N.S.; Bamfor, A.; Oppenheim, B.; et al. Seeking the source of Pseudomonas aeruginosa infections in a recently opened hospital: An observational study using whole-genome sequencing. BMJ Open 2014, 4, e006278. [Google Scholar] [CrossRef] [PubMed]
- Van Mansfeld, R.; Jongerden, I.; Bootsma, M.; Buiting, A.; Bonten, M.; Willems, R. The population genetics of Pseudomonas aeruginosa isolates from different patient populations exhibits high-level host specificity. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Raoult, D.; Fournier, P.E. Bacterial strain typing in the genomic era. FEMS Microbiol. Rev. 2009, 33, 892–916. [Google Scholar] [CrossRef] [PubMed]
- Van der Bij, A.K.; Van Mansfeld, R.; Peirano, G.; Goessens, W.H.F.; Severin, J.A.; Pitout, J.D.D.; Willems, R.; Van Westreenen, M. First outbreak of vim-2 metallo-β-lactamase-producing Pseudomonas aeruginosa in the netherlands: Microbiology, epidemiology and clinical outcomes. Int. J. Antimicrob. Agents 2011, 37, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Sobral, D.; Mariani-Kurkdjian, P.; Bingen, E.; Vu-Thien, H.; Hormigos, K.; Lebeau, B.; Loisy-Hamon, F.; Munck, A.; Vergnaud, G.; Pourcel, C. A new highly discriminatory multiplex capillary-based mlva assay as a tool for the epidemiological survey of Pseudomonas aeruginosa in cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2247–2256. [Google Scholar] [CrossRef] [PubMed]
- Jabalameli, F.; Mirsalehian, A.; Sotoudeh, N.; Jabalameli, L.; Aligholi, M.; Khoramian, B.; Taherikalani, M.; Emaneini, M. Multiple-locus variable number of tandem repeats (vntr) fingerprinting (mlvf) and antibacterial resistance profiles of extended spectrum beta lactamase (esbl) producing Pseudomonas aeruginosa among burnt patients in tehran. Burns 2011, 37, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Turton, J.F.; Turton, S.E.; Yearwood, L.; Yarde, S.; Kaufmann, M.E.; Pitt, T.L. Evaluation of a nine-locus variable-number tandem-repeat scheme for typing of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 2010, 16, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Vu-Thien, H.; Corbineau, G.; Hormigos, K.; Fauroux, B.; Corvol, H.; Clement, A.; Vergnaud, G.; Pourcel, C. Multiple-locus variable-number tandem-repeat analysis for longitudinal survey of sources of Pseudomonas aeruginosa infection in cystic fibrosis patients. J. Clin. Microbiol. 2007, 45, 3175–3183. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Mendis, N.; Trigui, H.; Oliver, J.D.; Faucher, S.P. The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 2014, 5, 258. [Google Scholar] [CrossRef] [PubMed]
- ASTM D5246-15. Standard Test Method for Isolation and Enumeration of Pseudomonas aeruginosa from Water; ASTM International: West Conshohocken, PA, USA, 2015; p. 5. [Google Scholar]
- Onteniente, L.; Brisse, S.; Tassios, P.T.; Vergnaud, G. Evaluation of the polymorphisms associated with tandem repeats for Pseudomonas aeruginosa strain typing. J. Clin. Microbiol. 2003, 41, 4991–4997. [Google Scholar] [CrossRef] [PubMed]
- Crivaro, V.; Di Popolo, A.; Caprio, A.; Lambiase, A.; Di Resta, M.; Borriello, T.; Scarcella, A.; Triassi, M.; Zarrilli, R. Pseudomonas aeruginosa in a neonatal intensive care unit: Molecular epidemiology and infection control measures. BMC Infect. Dis. 2009, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- Kotay, S.; Chai, W.; Guilford, W.; Barry, K.; Mathers, A.J. Spread from the sink to the patient: In situ study using green fluorescent protein (gfp)-expressing escherichia coli to model bacterial dispersion from hand-washing sink-trap reservoirs. Appl. Environ. Microbiol. 2017, 83, e03327-16. [Google Scholar] [CrossRef] [PubMed]
- Salm, F.; Deja, M.; Gastmeier, P.; Kola, A.; Hansen, S.; Behnke, M.; Gruhl, D.; Leistner, R. Prolonged outbreak of clonal mdr Pseudomonas aeruginosa on an intensive care unit: Contaminated sinks and contamination of ultra-filtrate bags as possible route of transmission? Antimicrob. Resist. Infect. Control 2016, 5, 53. [Google Scholar] [CrossRef] [PubMed]
- Bédard, E.; Prévost, M.; Déziel, E. Pseudomonas aeruginosa in premise plumbing of large buildings. MicrobiologyOpen 2016, 5, 937–956. [Google Scholar] [CrossRef] [PubMed]
- Dwidjosiswojo, Z.; Richard, J.; Moritz, M.M.; Dopp, E.; Flemming, H.-C.; Wingender, J. Influence of copper ions on the viability and cytotoxicity of Pseudomonas aeruginosa under conditions relevant to drinking water environments. Int. J. Hyg. Environ. Health 2011, 214, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Bédard, E.; Charron, D.; Lalancette, C.; Déziel, E.; Prévost, M. Recovery of Pseudomonas aeruginosa culturability following copper- and chlorine-induced stress. FEMS Microbiol. Lett. 2014, 356, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Blanc, D.S.; Nahimana, I.; Petignat, C.; Wenger, A.; Bille, J.; Francioli, P. Faucets as a reservoir of endemic Pseudomonas aeruginosa colonization/infections in intensive care units. Intensive Care Med. 2004, 30, 1964–1968. [Google Scholar] [CrossRef] [PubMed]
- Garvey, M.I.; Bradley, C.W.; Tracey, J.; Oppenheim, B. Continued transmission of Pseudomonas aeruginosa from a wash hand basin tap in a critical care unit. J. Hosp. Infec. 2016, 94, 8–12. [Google Scholar] [CrossRef] [PubMed]
- International Organization for Standardization (ISO). Water Quality—Detection and Enumeration of Pseudomonas aeruginosa—Method by Membrane Filtration; International Organization for Standardization: Geneva, Switzerland, 2006; p. 12. [Google Scholar]
- Lee, C.S.; Wetzel, K.; Buckley, T.; Wozniak, D.; Lee, J. Rapid and sensitive detection of Pseudomonas aeruginosa in chlorinated water and aerosols targeting gyrb gsene using real-time pcr. J. Appl. Microbiol. 2011, 111, 893–903. [Google Scholar] [CrossRef] [PubMed]
| Confirmed/Presumptive Pseudomonas aeruginosa (Pa) Isolates | |||||||
|---|---|---|---|---|---|---|---|
| (Confirmed/Presumptive Pa-Positive Environmental Sample) | |||||||
| Incubation Time (h) | Drain | Faucet | Surface | Total | |||
| Aa | PIa | Aa | PIa | Aa | PIa | ||
| 48 | 9/13 | 12/12 | — | — | 0/1 | 2/4 | 23/30 |
| (7/8) | (7/7) | — | — | (0/1) | (2/2) | (10/12) 1 | |
| 72 | 2/2 | 2/2 | — | — | — | 0/1 | 4/5 |
| (1/1) | (1/1) | — | — | — | (0/1) | (1/2) 1 | |
| 96 | — | — | — | 3/4 | 1/1 | — | 4/5 |
| — | — | — | (1/2) | (1/1) | — | (2/3) | |
| 240 | — | — | — | 4/4 | — | 3/3 | 7/7 |
| — | — | — | (2/2) | — | (2/2) | (4/4) | |
| Total | 11/15 | 14/14 | — | 7/8 | 1/2 | 5/8 | 38/47 |
| (8/9) | (8/8) | — | (3/4) | (1/2) | (4/5) | (17/21) | |
| Hospital | Incubation Time (h) | % Pa-Positive (n Sampled Sites) | ||
|---|---|---|---|---|
| Drain | Faucet | Surface | ||
| A | Up to 240 | 33 (18) | 17 (18) | 22 (18) |
| 48 1 | 28 (18) | 0 (18) | 6 (18) | |
| B to E | 48 1 | 51 (210) | 1 (210) | 10 (60) |
| MS142 | MS211 | MS213 | MS215 | MS216 | MS222 | MS223 | |
|---|---|---|---|---|---|---|---|
| All strains (n = 70) | |||||||
| No. of observed alleles | 6 | 7 | 7 | 6 | 3 | 6 | 5 |
| Alleles | 1–5; 7 | 2–8 | 1–5; 8; 9 | 1–6 | 1–2; 4 | 1–6 | 2–5; 7 |
| Typability (%) | 96 | 89 | 90 | 86 | 96 | 82 | 86 |
| HGDI | 0.72 | 0.83 | 0.72 | 0.75 | 0.49 | 0.74 | 0.60 |
| All environmental strains (n = 38) | |||||||
| No. of observed alleles | 4 | 4 | 3 | 4 | 2 | 2 | 3 |
| Alleles | 1–2; 4–5 | 2–4; 6 | 1; 4–5 | 1–2; 4; 6 | 1–2 | 2; 4 | 2–4 |
| Typability (%) | 100 | 97 | 100 | 100 | 95 | 84 | 92 |
| All clinical strains (n = 32) | |||||||
| No. of observed alleles | 6 | 7 | 7 | 6 | 3 | 6 | 5 |
| Alleles | 1–5; 7 | 2–8 | 1–5; 8; 9 | 1–6 | 1–2; 4 | 1–6 | 2–5; 7 |
| Typability (%) | 91 | 79 | 79 | 70 | 97 | 79 | 79 |
| Youenoue et al., 2014 (n = 62) | |||||||
| No. of observed alleles | 9 | 9 | 11 | 8 | 5 | 7 | 6 |
| Alleles | 1–7; 12 | 2–9 | 0–7; 9–10 | 1–7 | 1–5 | 1–5; 7 | 2–7 |
| Typability (%) | 100 | 97 | 98 | 97 | 98 | 99 | 100 |
| HGDI | 0.83 | 0.79 | 0.86 | 0.86 | 0.65 | 0.76 | 0.70 |
| Vu-Thien et al., 2007 (n = 24) | |||||||
| No. of observed alleles | 9 | 8 | 7 | 7 | 4 | 7 | 7 |
| Alleles | 1–7 | 2–8 | 3–7; 9 | 1–6 | 1–4 | 1–6 | 2–7 |
| Typability (%) | 100 | 100 | 96 | 100 | 96 | 100 | 75 |
| HGDI | 0.81 | 0.76 | 0.85 | 0.80 | 0.64 | 0.76 | 0.77 |
| % Paired Pseudomonas aeruginosa Positivity at a Sink (Odds Ratio) | |||||
| n Positive | Drain | Faucet | Sink Surface | ||
| Hospitals A & B | Drain | 108 | — | 7 (3.3) | 20 (1.3) |
| Faucet | 3 | 67 | — | 40 (6) | |
| Sink surface | 9 | 60 | 20 | — | |
| n Positive | Drain | Faucet | Water | ||
| Hospitals B, C, D & E | Drain | 108 | — | 7 (3.3) | 20 (1.3) |
| Faucet | 3 | 67 | — | 40 (6) | |
| Water | 9 | 60 | 20 | — | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lalancette, C.; Charron, D.; Laferrière, C.; Dolcé, P.; Déziel, E.; Prévost, M.; Bédard, E. Hospital Drains as Reservoirs of Pseudomonas aeruginosa: Multiple-Locus Variable-Number of Tandem Repeats Analysis Genotypes Recovered from Faucets, Sink Surfaces and Patients. Pathogens 2017, 6, 36. https://doi.org/10.3390/pathogens6030036
Lalancette C, Charron D, Laferrière C, Dolcé P, Déziel E, Prévost M, Bédard E. Hospital Drains as Reservoirs of Pseudomonas aeruginosa: Multiple-Locus Variable-Number of Tandem Repeats Analysis Genotypes Recovered from Faucets, Sink Surfaces and Patients. Pathogens. 2017; 6(3):36. https://doi.org/10.3390/pathogens6030036
Chicago/Turabian StyleLalancette, Cindy, Dominique Charron, Céline Laferrière, Patrick Dolcé, Eric Déziel, Michèle Prévost, and Emilie Bédard. 2017. "Hospital Drains as Reservoirs of Pseudomonas aeruginosa: Multiple-Locus Variable-Number of Tandem Repeats Analysis Genotypes Recovered from Faucets, Sink Surfaces and Patients" Pathogens 6, no. 3: 36. https://doi.org/10.3390/pathogens6030036






