Evolution and Divergence of H3N8 Equine Influenza Viruses Circulating in the United Kingdom from 2013 to 2015
Abstract
:1. Introduction
2. Results
2.1. Genetic Analyses—HA
2.2. Genetic Analyses—NA
2.3. Antigenic Characterisation
3. Discussion
4. Materials and Methods
4.1. Diagnostic Testing for the Presence of EIV
4.2. Virus Isolation in Eggs
4.3. RNA Extraction, RT-PCR and Sequencing
4.4. Sequence Analysis and Phylogenetic Trees
4.5. Haemagglutination Inhibition (HI) Assay
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Weis, W; Brown, J.H.; Cusack, S.; Paulson, J.C.; Skehel, J.J.; Wiley, D.C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 1988, 333, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.D. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 2004, 78, 12665–12667. [Google Scholar] [CrossRef] [PubMed]
- Halbherr, S.J.; Ludersdorfer, T.H.; Ricklin, M.; Locher, S.; Berger Rentsch, M.; Summerfield, A.; Zimmer, G. Biological and protective properties of immune sera directed to influenza virus neuraminidase. J. Virol. 2015, 89, 1550–1563. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, T.; Tsujimura, K.; Kondo, T.; Hobo, S.; Matsumura, T. Efficacy of oseltamivir phosphate to horses inoculated with equine influenza A virus. J. Vet. Med. Sci. 2006, 68, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, T.; Bannai, H.; Nemoto, M.; Tsujimura, K.; Kondo, T.; Muranaka, M.; Hobo, S.; Minamijima, Y.H.; Yamada, M.; Matsumura, T. Efficacy of a single intravenous dose of the neuraminidase inhibitor peramivir in the treatment of equine influenza. Vet. J. 2012, 193, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Waddell, G.H.; Teigland, M.B.; Sigel, M.M. A new influenza virus associated with equine respiratory disease. J. Am. Vet. Med. Assoc. 1963, 143, 587–590. [Google Scholar] [PubMed]
- Bryant, N.A.; Rash, A.S.; Russell, C.A.; Ross, J.; Cooke, A.; Bowman, S.; MacRae, S.; Lewis, N.S.; Paillot, R.; Zanoni, R.; et al. Antigenic and genetic variations in European and North American equine influenza virus strains (H3N8) isolated from 2006 to 2007. Vet. Microbiol. 2009, 138, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.L.; Rash, A.S.; Blinman, D.; Bowman, S.; Chambers, T.M.; Daly, J.M.; Damiani, A.; Joseph, S.; Lewis, N.; McCauley, J.W.; et al. Development of a surveillance scheme for equine influenza in the UK and characterisation of viruses isolated in Europe, Dubai and the USA from 2010 to 2012. Vet. Microbiol. 2014, 169, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Burrows, R.; Denyer, M.; Goodrige, D.; Hamilton, F. Field and laboratory studies of equine influenza viruses isolated in 1979. Vet. Rec. 1981, 109, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Cowled, B.; Ward, M.P.; Hamilton, S.; Garner, G. The equine influenza epidemic in Australia: Spatial and temporal descriptive analyses of a large propagating epidemic. Prev. Vet. Med. 2009, 92, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Livesay, G.J.; O’Neill, T.; Hannant, D.; Yadav, M.P.; Mumford, J.A. The outbreak of equine influenza (H3N8) in the United Kingdom in 1989: Diagnostic use of an antigen capture ELISA. Vet. Rec. 1993, 133, 515–519. [Google Scholar] [CrossRef]
- Ito, M.; Nagai, M.; Hayakawa, Y.; Komae, H.; Murakami, N.; Yotsuya, S.; Asakura, S.; Sakoda, Y.; Kida, H. Genetic Analyses of an H3N8 Influenza Virus Isolate, Causative Strain of the Outbreak of Equine Influenza at the Kanazawa Racecourse in Japan in 2007. J. Vet. Med. Sci. 2008, 70, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Newton, J.R.; Daly, J.M.; Spencer, L.; Mumford, J.A. Description of the outbreak of equine influenza (H3N8) in the United Kingdom in 2003, during which recently vaccinated horses in Newmarket developed respiratory disease. Vet. Rec. 2006, 158, 185–192. [Google Scholar] [CrossRef]
- Virmani, N.; Bera, B.C.; Singh, B.K.; Shanmugasundaram, K.; Gulati, B.R.; Barua, S.; Vaid, R.K.; Gupta, A.K.; Singh, R.K. Equine influenza outbreak in India (2008–09): Virus isolation, sero-epidemiology and phylogenetic analysis of HA gene. Vet. Microbiol. 2010, 143, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Alves Beuttemmüller, E.; Woodward, A.; Rash, A.; Dos Santos Ferraz, L.E.; Fernandes Alfieri, A.; Alfieri, A.A.; Elton, D. Characterisation of the epidemic strain of H3N8 equine influenza virus responsible for outbreaks in South America in 2012. Virol. J. 2016, 19, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Perglione, C.O.; Gildea, S.; Rimondi, A.; Miño, S.; Vissani, A.; Carossino, M.; Cullinane, A.; Barrandeguy, M. Epidemiological and virological findings during multiple outbreaks of equine influenza in South America in 2012. Influenza Other Respir. Viruses 2016, 10, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, T.; Niwa, H.; Tsujimura, K.; Kondo, T.; Matsumura, T. Epidemic of equine influenza among vaccinated racehorses in Japan in 2007. J. Vet. Med. Sci. 2008, 70, 623–625. [Google Scholar] [CrossRef] [PubMed]
- Worobey, M.; Han, G.Z.; Rambaut, A. A synchronised global sweep of the internal genes of modern avian influenza virus. Nature 2014, 508, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.C.; Chambers, T.M.; Holland, R.E., Jr.; Morley, P.S.; Haines, D.M.; Townsend, H.G.; Barrandeguy, M. Diverged evolution of recent equine-2 influenza (H3N8) viruses in the Western Hemisphere. Arch. Virol. 2001, 146, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Bernardino, P.N.; Mapes, S.M.; Corbin, R.; Pusterla, N. Pyrosequencing as a fast and reliable tool to determine clade affiliation for equine Influenza A virus. J. Vet. Diagn. Investig. 2016, 28, 323–326. [Google Scholar] [CrossRef] [PubMed]
- Gildea, S.; Quinlivan, M.; Arkins, S.; Cullinane, A. The molecular epidemiology of equine influenza in Ireland from 2007 to 2010 and its international significance. Equine Vet. J. 2012, 44, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Gildea, S.; Fitzpatrick, D.A.; Cullinane, A. Epidemiological and virological investigations of equine influenza outbreaks in Ireland (2010–2012). Influenza Other Respir. Viruses 2013, 7 (Suppl. 4), 61–72. [Google Scholar] [CrossRef] [PubMed]
- Legrand, L.J.; Pitel, P.H.; Cullinane, A.A.; Fortier, G.D.; Pronost, S.L. Genetic evolution of equine influenza strains isolated in France from 2005 to 2010. Equine Vet. J. 2015, 47, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Guo, W.; Huang, W.Q.; Li, H.M.; Zhao, L.P.; Dai, L.L.; He, N.; Hao, X.F.; Xiang, W.H. Genetic evolution of equine influenza viruses isolated in China. Arch. Virol. 2010, 155, 1425–1432. [Google Scholar] [CrossRef] [PubMed]
- Yondon, M.; Heil, G.L.; Burks, J.P.; Zayat, B.; Waltzek, T.B.; Jamiyan, B.O.; McKenzie, P.P.; Krueger, W.S.; Friary, J.A.; Gray, G.C. Isolation and characterization of H3N8 equine influenza A virus associated with the 2011 epizootic in Mongolia. Influenza Other Respir. Viruses 2013, 7, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Binns, M.M.; Daly, J.M.; Chirnside, E.D.; Mumford, J.A.; Wood, J.M.; Richards, C.M.; Daniels, R.S. Genetic and antigenic analysis of an equine influenza H 3 isolate from the 1989 epidemic. Arch. Virol. 1993, 130, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Barbic, L.; Madic, J.; Turk, N.; Daly, J. Vaccine failure caused an outbreak of equine influenza in Croatia. Vet. Microbiol. 2009, 133, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Cullinane, A.; Elton, D.; Mumford, J. Equine influenza—Surveillance and control. Influenza Other Respir. Viruses 2010, 4, 339–344. [Google Scholar] [CrossRef] [PubMed]
- The World Organisation for Animal Health (OIE). Expert surveillance panel on equine influenza vaccine composition—Conclusions and recommendations. Off. Int. Epizoot. Bull. 2016, 2, 61–63. [Google Scholar]
- GISAID EpiFlu Database. Editorial: Action stations: The time for sitting on flu data is over. Nature 2006, 441, 1028. [Google Scholar]
- Collins, P.J.; Vachieri, S.G.; Haire, L.F.; Ogrodowicz, R.W.; Martin, S.R.; Walker, P.A.; Xiong, X.; Gamblin, S.J.; Skehel, J.J. Recent evolution of equine influenza and the origin of canine influenza. Proc. Natl. Acad. Sci. USA 2014, 111, 11175–11180. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Haire, L.F.; Stevens, D.J.; Collins, P.J.; Lin, Y.P.; Blackburn, G.M.; Hay, A.J.; Gamblin, S.J.; Skehel, J.J. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 2006, 443, 45–49. [Google Scholar] [CrossRef] [PubMed]
- The World Organisation for Animal Health (OIE). Expert surveillance panel on equine influenza vaccine composition—Conclusions and recommendations. Off. Int. Epizoot. Bull. 2010, 2, 44–45. [Google Scholar]
- Bryant, N.A.; Rash, A.S.; Woodward, A.L.; Medcalf, E.; Helwegen, M.; Wohlfender, F.; Cruz, F.; Herrmann, C.; Borchers, K.; Tiwari, A.; et al. Isolation and characterisation of equine influenza viruses (H3N8) from Europe and North America from 2008 to 2009. Vet. Microbiol. 2011, 147, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Corti, D.; Suguitan, A.L., Jr.; Pinna, D.; Silacci, C.; Fernandez-Rodriguez, B.M.; Vanzetta, F.; Santos, C.; Luke, C.J.; Torres-Velez, F.J.; Temperton, N.J.; et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Investig. 2010, 120, 1663–1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dreyfus, C.; Ekiert, D.C.; Wilson, I.A. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J. Virol. 2013, 87, 7149–7154. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, T.; Cullinane, A.; Gildea, S.; Bannai, H.; Nemoto, M.; Tsujimura, K.; Kondo, T.; Matsumura, T. The potential impact of a single amino-acid substitution on the efficacy of equine influenza vaccines. Equine Vet. J. 2015, 47, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, T.; Nemoto, M.; Bannai, H.; Tsujimura, K.; Kondo, T.; Matsumura, T.; Gildea, S.; Cullinane, A. Assessment of antigenic difference of equine influenza virus strains by challenge study in horses. Influenza Other Respir. Viruses 2016, 10, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Cook, R.F.; Sinclair, R.; Mumford, J.A. Detection of influenza nucleoprotein antigen in nasal secretions from horses infected with A/equine influenza (H3N8) viruses. J. Virol. Methods 1988, 20, 1–12. [Google Scholar] [CrossRef]
- OIE. Chapter 2.5.7 Equine influenza. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; OIE: Paris, France, 2016; pp. 1–16. Available online: http://www.oie.int/international-standard-setting/terrestrial-manual/access-online/ (accessed on 3 November 2016).
- Rash, A.; Woodward, A.; Bryant, N.; McCauley, J.; Elton, D. An efficient genome sequencing method for equine influenza [H3N8] virus reveals a new polymorphism in the PA-X protein. Virol. J. 2014, 159. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefor, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]

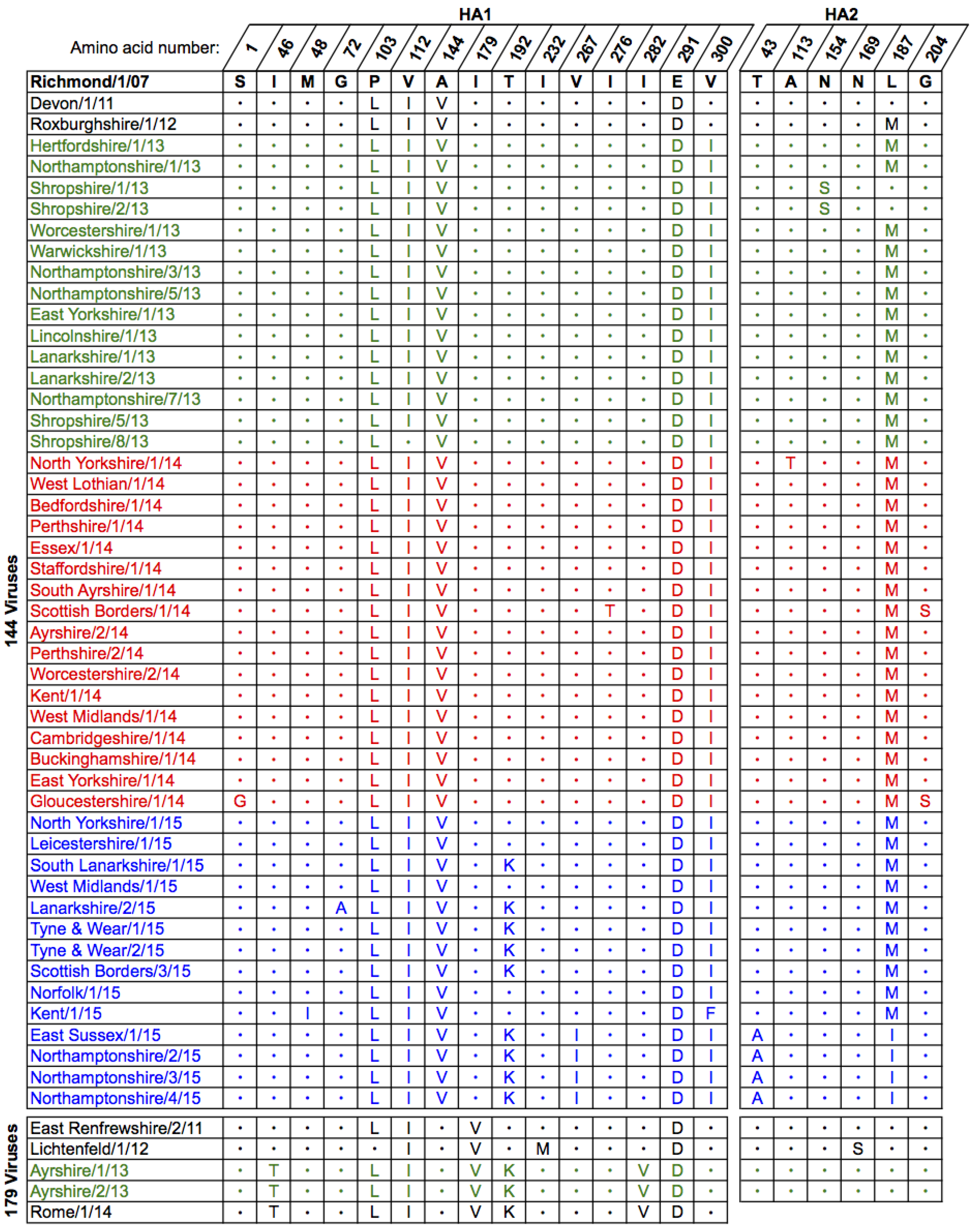
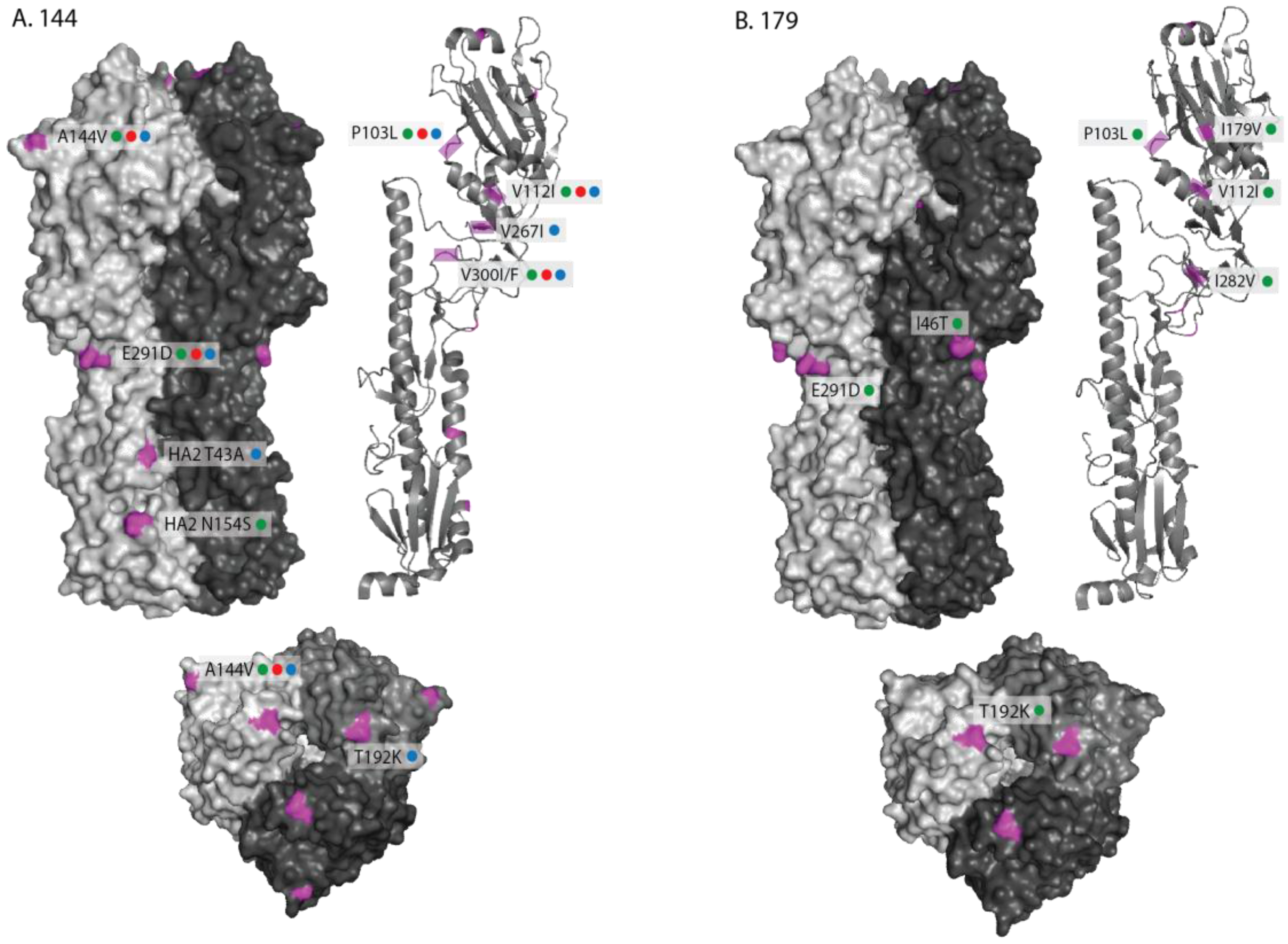
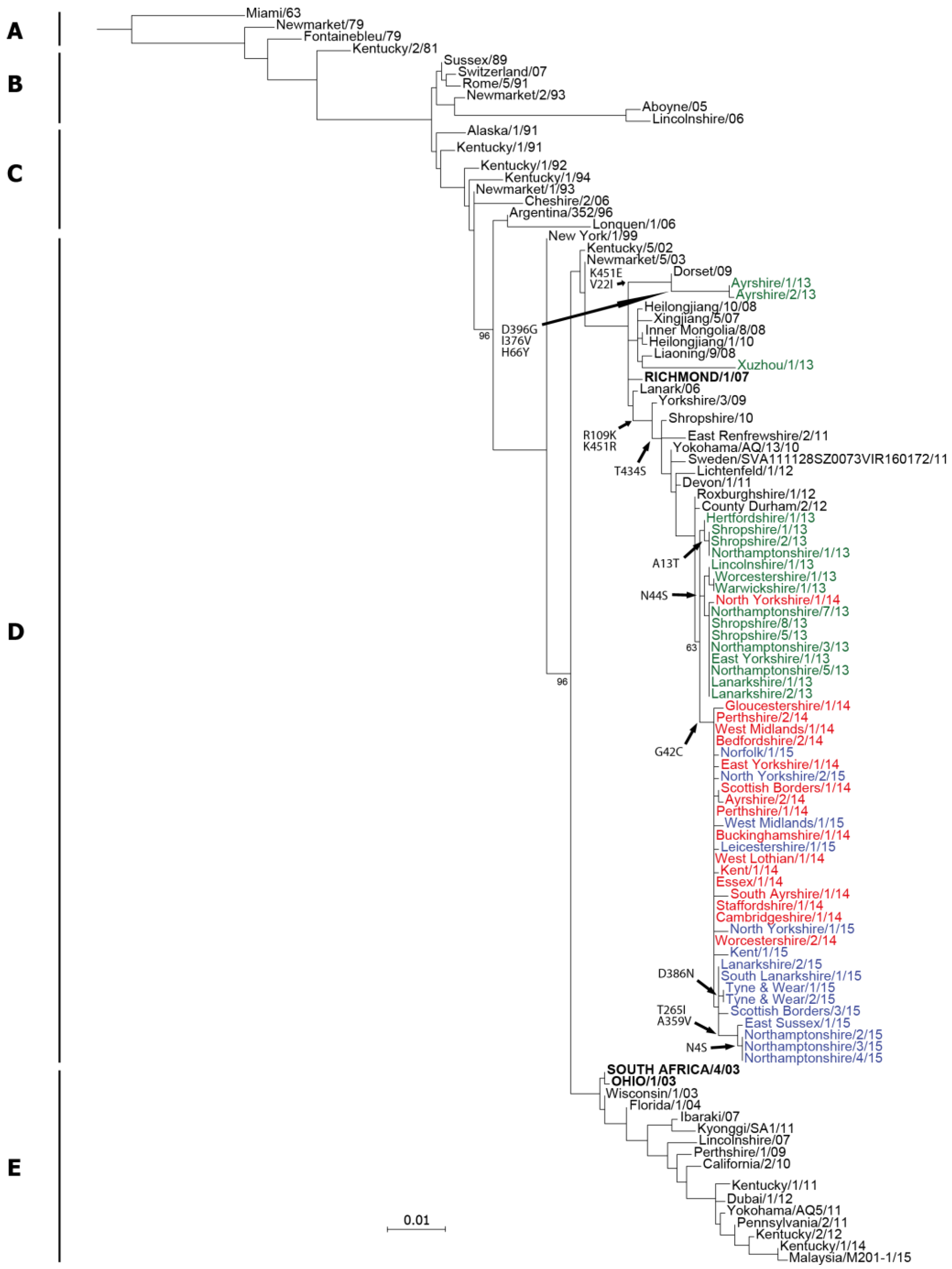

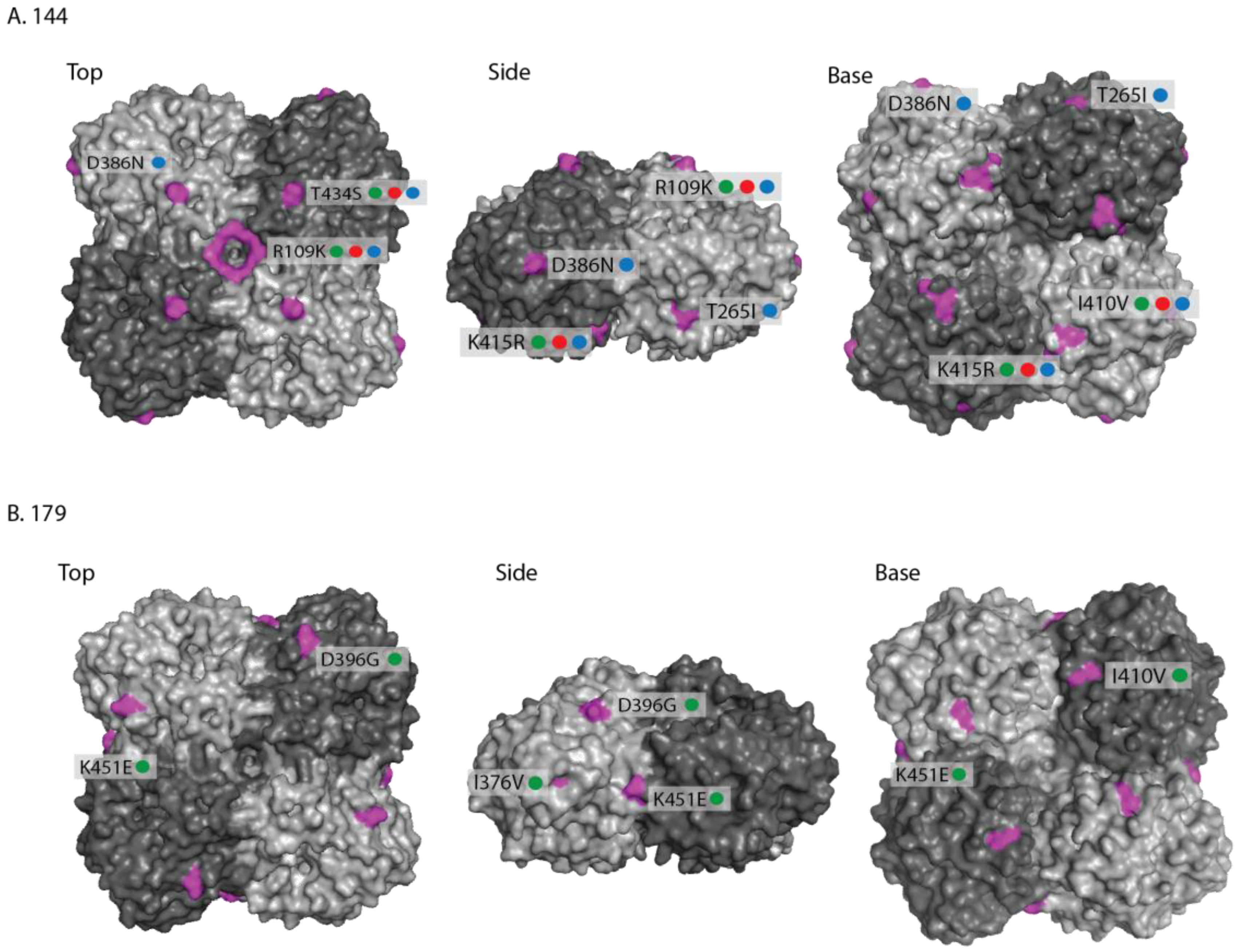
| Date | Location (county) | Country | Detection method | Isolate name(s) | HA acc. | NA acc. |
|---|---|---|---|---|---|---|
| 2013 | ||||||
| Feb | Ayrshire | Scotland | qRT-PCR | Ayrshire/1/13 | EPI492691 | EPI497568 |
| Ayrshire/2/13 | EPI838696 | EPI838697 | ||||
| Jun | Hertfordshire | England | qRT-PCR | Hertfordshire/1/13 | EPI492692 | EPI497569 |
| Jul | Northamptonshire | England | qRT-PCR | Northamptonshire/1/13 | EPI492817 | EPI493625 |
| Jul | Shropshire | England | qRT-PCR | Shropshire/1/13 | EPI838698 | EPI838699 |
| Shropshire/2/13 | EPI838700 | EPI838701 | ||||
| Sep | Worcestershire | England | qRT-PCR | Worcestershire/1/13 | EPI492821 | EPI497570 |
| Sep | Warwickshire | England | qRT-PCR | Warwickshire/1/13 | EPI492822 | EPI497571 |
| Sep | Northamptonshire | England | qRT-PCR | Northamptonshire/3/13 | EPI838702 | EPI838703 |
| Northamptonshire/5/13 | EPI838916 | EPI838917 | ||||
| Sep | East Yorkshire | England | qRT-PCR | East Yorkshire/1/13 | EPI492823 | EPI497575 |
| Sep | Lincolnshire | England | qRT-PCR | Lincolnshire/1/13 | EPI838918 | EPI838919 |
| Sep | Lanarkshire | Scotland | qRT-PCR | Lanarkshire/1/13 | EPI493612 | EPI493613 |
| Oct | Northamptonshire | England | qRT-PCR | Northamptonshire/7/13 | EPI493616 | EPI493617 |
| Oct | Shropshire | England | qRT-PCR | Shropshire/5/13 | EPI493619 | EPI493620 |
| Dec | Shropshire | England | qRT-PCR | Shropshire/8/13 | EPI493622 | EPI493623 |
| 2014 | ||||||
| Mar | North Yorkshire | England | qRT-PCR | North Yorkshire/1/14 | EPI821293 | EPI821294 |
| Aug | West Lothian | Scotland | qRT-PCR | West Lothian/1/14 | EPI839642 | EPI839656 |
| Aug | Bedfordshire | England | qRT-PCR | Bedfordshire/2/14 | EPI821287 | EPI821288 |
| Sep | Perthshire | Scotland | qRT-PCR | Perthshire/1/14 | EPI839711 | EPI839712 |
| Sep | Essex | England | qRT-PCR | Essex/1/14 | EPI821295 | EPI821296 |
| Sep | Staffordshire | England | qRT-PCR | Staffordshire/1/14 | EPI839713 | EPI839714 |
| Oct | South Ayrshire | Scotland | qRT-PCR | South Ayrshire/1/14 | EPI821285 | EPI821286 |
| Oct | Scottish Borders | Scotland | qRT-PCR | Scottish Borders/1/14 | EPI839715 | EPI839716 |
| Oct | Ayrshire | Scotland | qRT-PCR | Ayrshire/2/14 | EPI821145 | EPI821202 |
| Oct | Perthshire | Scotland | qRT-PCR | Perthshire/2/14 | EPI839717 | EPI839718 |
| Oct | Worcestershire | England | qRT-PCR | Worcestershire/2/14 | EPI839719 | EPI839720 |
| Oct | Kent | England | qRT-PCR | Kent/1/14 | EPI839721 | EPI839722 |
| Oct | West Midlands | England | NP ELISA | West Midlands/1/14 | EPI839723 | EPI839724 |
| Nov | Cambridgeshire | England | qRT-PCR | Cambridgeshire/1/14 | EPI821289 | EPI821290 |
| Nov | Buckinghamshire | England | qRT-PCR | Buckinghamshire/1/14 | EPI651398 | EPI651400 |
| Nov | East Yorkshire | England | qRT-PCR | East Yorkshire/1/14 | EPI821291 | EPI821292 |
| Nov | Gloucestershire | England | qRT-PCR | Gloucestershire/1/14 | EPI821297 | EPI821298 |
| 2015 | ||||||
| Mar | North Yorkshire | England | qRT-PCR | North Yorkshire/1/15 | EPI686738 | EPI686739 |
| Apr | Leicestershire | England | qRT-PCR | Leicestershire/1/15 | EPI687475 | EPI694842 |
| Jun | South Lanarkshire | Scotland | qRT-PCR | South Lanarkshire/1/15 | EPI643192 | EPI643193 |
| Jul | West Midlands | England | qRT-PCR | West Midlands/1/15 | EPI650051 | EPI650052 |
| Jul | Lanarkshire | Scotland | qRT-PCR | Lanarkshire/2/15 | EPI650053 | EPI650054 |
| Jul | Tyne & Wear | England | qRT-PCR | Tyne & Wear/1/15 | EPI650056 | EPI651799 |
| Tyne & Wear/2/15 | EPI651395 | EPI650055 | ||||
| Jul | Scottish Borders | Scotland | qRT-PCR | Scottish Borders/3/15 | EPI710985 | EPI710986 |
| Aug | Norfolk | England | qRT-PCR | Norfolk/1/15 | EPI686736 | EPI686737 |
| Oct | Kent | England | qRT-PCR | Kent/1/15 | EPI673530 | EPI673531 |
| Nov | East Sussex | England | qRT-PCR | East Sussex/1/15 | EPI673532 | EPI673533 |
| Dec | Northamptonshire | England | qRT-PCR | Northamptonshire/2/15 | EPI686740 | EPI839779 |
| Northamptonshire/3/15 | EPI839794 | EPI839809 | ||||
| Northamptonshire/4/15 | EPI839821 | EPI839822 | ||||
| Reference ferret antisera | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| New/1/93 | New/2/93 | Rich/1/07 | Dev/1/11 | ER/2/11 | Ayr/1/13 | SA/4/03 | Dor/09 | RGdS/1/12 | Ky/1/14 | |
| Am | Eu | FC2 | FC2 | FC2 | FC2 | FC1 | FC1 | FC1 | FC1 | |
| Reference strains | ||||||||||
| Newmarket/1/93 | 362 | 11 | 362 | 181 | 256 | 362 | 32 | 64 | 32 | 64 |
| Newmarket/2/93 | 91 | 512 | 64 | 128 | 64 | 128 | <8 | 32 | 8 | 23 |
| Richmond/1/07 | 181 | 32 | 256 | 91 | 128 | 256 | 64 | 128 | 128 | 128 |
| Devon/1/11 | 362 | 64 | 724 | 512 | 362 | 512 | 181 | 256 | 128 | 256 |
| East Renfrewshire/2/11 | 256 | 32 | 724 | 256 | 256 | 512 | 128 | 256 | 128 | 256 |
| Ayrshire/1/13 | 256 | 16 | 512 | 256 | 128 | 724 | 64 | 128 | 45 | 256 |
| South Africa/4/03 | 23 | 8 | 91 | 64 | 45 | 181 | 256 | 362 | 256 | 512 |
| Dorset/09 | 64 | 32 | 181 | 91 | 91 | 256 | 724 | 1024 | 1024 | 2048 |
| Rio Grande do Sul/1/12 | 128 | 23 | 256 | 128 | 64 | 256 | 724 | 1024 | 1024 | 2048 |
| Kentucky/1/14 | 64 | 16 | 128 | 128 | 64 | 128 | 1024 | 2048 | 2896 | 2896 |
| 2013-15 isolates | ||||||||||
| Ayrshire/2/13 | 362 | 16 | 724 | 181 | 128 | 724 | 128 | 256 | 91 | 362 |
| Northamptonshire/1/13 | 724 | 91 | 1024 | 724 | 362 | 724 | 128 | 256 | 181 | 256 |
| Shropshire/1/13 | 362 | 32 | 512 | 362 | 128 | 256 | 91 | 256 | 128 | 256 |
| Northamptonshire/3/13 | 362 | 64 | 1024 | 512 | 256 | 512 | 128 | 256 | 181 | 256 |
| Shropshire/8/13 | 256 | 32 | 512 | 362 | 128 | 256 | 128 | 191 | 128 | 256 |
| North Yorkshire/1/14 | 512 | 64 | 1024 | 512 | 181 | 512 | 64 | 362 | 181 | 256 |
| Scottish Borders/1/14 | 256 | 64 | 1024 | 512 | 256 | 512 | 128 | 256 | 128 | 256 |
| Perthshire/2/14 | 256 | 32 | 724 | 362 | 128 | 256 | 128 | 256 | 64 | 256 |
| Kent/1/14 | 256 | 32 | 724 | 256 | 128 | 256 | 128 | 256 | 128 | 256 |
| Buckinghamshire/1/14 | 362 | 32 | 1024 | 256 | 91 | 362 | 91 | 256 | 91 | 256 |
| Gloucestershire/1/14 | 362 | 45 | 1024 | 512 | 181 | 512 | 128 | 256 | 128 | 256 |
| South Lanarkshire/1/15 | 256 | 16 | 362 | 256 | 181 | 512 | 64 | 128 | 64 | 256 |
| Lanarkshire/2/15 | 256 | 16 | 362 | 128 | 91 | 362 | 64 | 128 | 64 | 256 |
| Scottish Borders/3/15 | 512 | 32 | 512 | 256 | 128 | 512 | 64 | 181 | 64 | 362 |
| Kent/1/15 | 256 | 32 | 512 | 256 | 128 | 256 | 128 | 256 | 91 | 256 |
| East Sussex/1/15 | 181 | 23 | 362 | 181 | 64 | 256 | 32 | 91 | 45 | 91 |
| Northamptonshire/2/15 | 512 | 23 | 362 | 362 | 91 | 724 | 64 | 128 | 32 | 256 |
| Northamptonshire/3/15 | 362 | 16 | 256 | 362 | 128 | 512 | 64 | 128 | 64 | 256 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rash, A.; Morton, R.; Woodward, A.; Maes, O.; McCauley, J.; Bryant, N.; Elton, D. Evolution and Divergence of H3N8 Equine Influenza Viruses Circulating in the United Kingdom from 2013 to 2015. Pathogens 2017, 6, 6. https://doi.org/10.3390/pathogens6010006
Rash A, Morton R, Woodward A, Maes O, McCauley J, Bryant N, Elton D. Evolution and Divergence of H3N8 Equine Influenza Viruses Circulating in the United Kingdom from 2013 to 2015. Pathogens. 2017; 6(1):6. https://doi.org/10.3390/pathogens6010006
Chicago/Turabian StyleRash, Adam, Rachel Morton, Alana Woodward, Olivia Maes, John McCauley, Neil Bryant, and Debra Elton. 2017. "Evolution and Divergence of H3N8 Equine Influenza Viruses Circulating in the United Kingdom from 2013 to 2015" Pathogens 6, no. 1: 6. https://doi.org/10.3390/pathogens6010006





