Mycobacterial Interspersed Repeat Unit–Variable Number Tandem Repeat Typing of Mycobacterium avium Strains Isolated from the Lymph Nodes of Free-Living Carnivorous Animals in Poland
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Culture
2.3. DNA Isolation
2.4. Strain Identification
2.5. IS901, IS900, and IS1245 Identification
2.6. MIRU-VNTR Identification
3. Results
3.1. Post-Mortem Examination
3.2. Mycobacterial Analysis and Species Designation
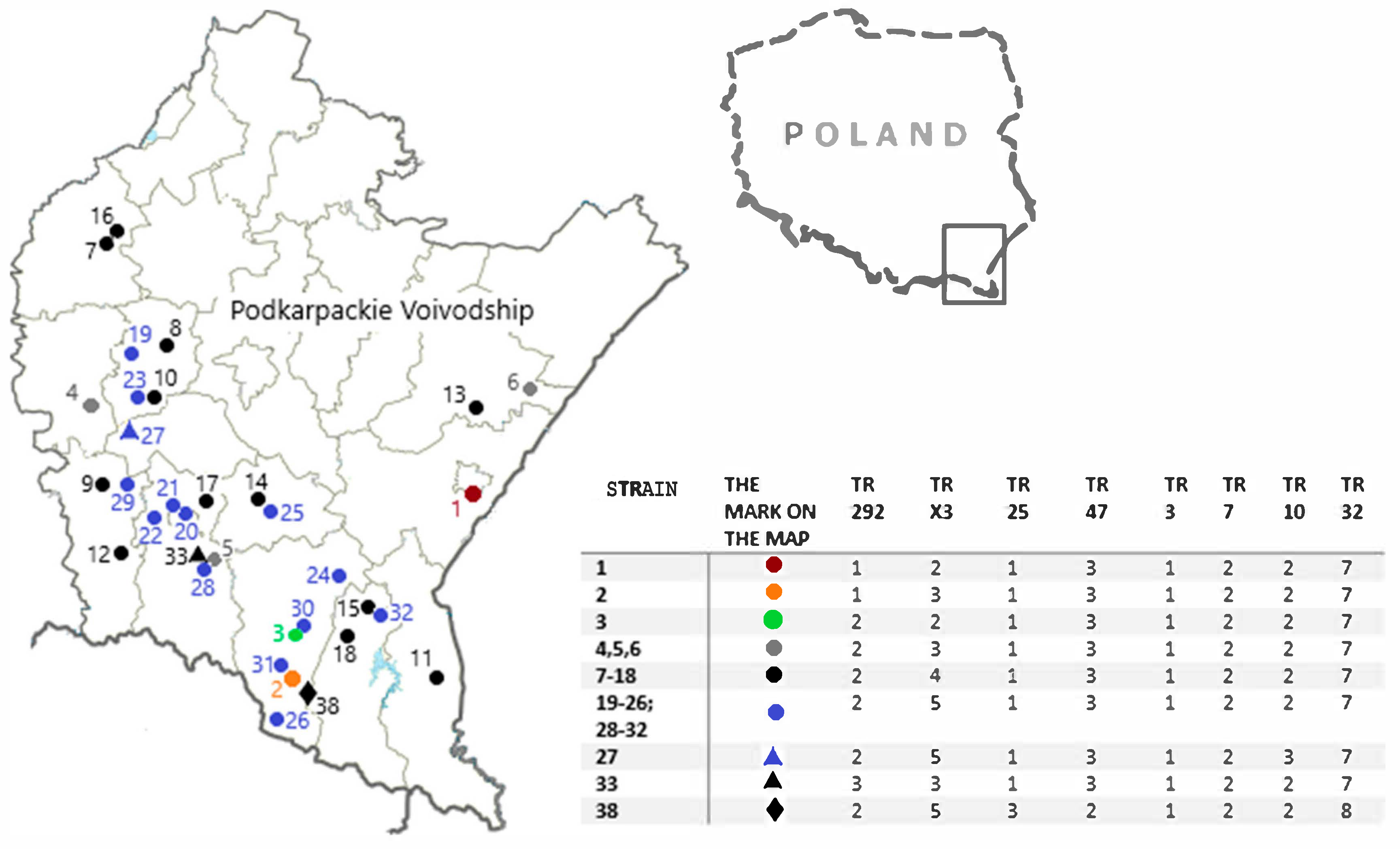
3.3. MIRU-VNTR Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Falkinham, J.O., 3rd. Current Epidemiologic Trends of the Nontuberculous Mycobacteria (NTM). Curr. Environ. Health Rep. 2016, 3, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Ghielmetti, G.; Giger, U. Mycobacterium avium: An Emerging Pathogen for Dog Breeds with Hereditary Immunodeficiencies. Curr. Clin. Microbiol. Rep. 2020, 7, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Johansen, M.D.; Herrmann, J.L.; Kremer, L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat. Rev. Microbiol. 2020, 18, 392–407. [Google Scholar] [CrossRef]
- Peterhans, S.; Landolt, P.; Friedel, U.; Oberhänsli, F.; Dennler, M.; Willi, B.; Senn, M.; Hinden, S.; Kull, K.; Kipar, A.; et al. Mycobacterium microti: Not Just a Coincidental Pathogen for Cats. Front. Vet. Sci. 2020, 7, 590037. [Google Scholar] [CrossRef] [PubMed]
- To, K.; Cao, R.; Yegiazaryan, A.; Owens, J.; Venketaraman, V. General Overview of Nontuberculous Mycobacteria Opportunistic Pathogens: Mycobacterium avium and Mycobacterium abscessus. J. Clin. Med. 2020, 9, 2541. [Google Scholar] [CrossRef]
- Cole, A.L.; Kirk, N.M.; Wang, L.; Hung, C.C.; Samuelson, J.P. Mycobacterium fortuitum abortion in a sow. J. Vet. Diagn. Investig. 2022, 34, 116–120. [Google Scholar] [CrossRef]
- Biet, F.; Boschiroli, M.L. Non-tuberculous mycobacterial infections of veterinary relevance. Res. Vet. Sci. 2014, 97, S69–S77. [Google Scholar] [CrossRef]
- van Ingen, J.; Turenne, C.Y.; Tortoli, E.; Wallace, R.J., Jr.; Brown-Elliott, B.A. A definition of the Mycobacterium avium complex for taxonomical and clinical purposes, a review. Int. J. Syst. Evol. Microbiol. 2018, 68, 3666–3677. [Google Scholar] [CrossRef]
- Mizzi, R.; Plain, K.M.; Whittington, R.; Timms, V.J. Global Phylogeny of Mycobacterium avium and Identification of Mutation Hotspots During Niche Adaptation. Front. Microbiol. 2022, 13, 892333. [Google Scholar] [CrossRef]
- Johnson, M.M.; Odell, J.A. Nontuberculous mycobacterial pulmonary infections. J. Thorac. Dis. 2014, 6, 210–220. [Google Scholar] [CrossRef]
- Gcebe, N.; Hlokwe, T.M. Non-tuberculous Mycobacteria in South African Wildlife: Neglected Pathogens and Potential Impediments for Bovine Tuberculosis Diagnosis. Front. Cell. Infect. Microbiol. 2017, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Didkowska, A.; Krajewska-Wędzina, M.; Klich, D.; Prolejko, K.; Orłowska, B.; Anusz, K. The Risk of False-Positive Serological Results for Paratuberculosis in Mycobacterium bovis-Infected Cattle. Pathogens 2021, 10, 1054. [Google Scholar] [CrossRef] [PubMed]
- van Ingen, J.; Griffith, D.E.; Aksamit, T.R.; Wagner, D. Pulmonary diseases caused by non-tuberculous mycobacteria. Eur. Respir. Monogr. 2012, 58, 25–37. [Google Scholar] [CrossRef]
- Slany, M.; Ulmann, V.; Slana, I. Avian Mycobacteriosis: Still Existing Threat to Humans. BioMed Res. Int. 2016, 2016, 4387461. [Google Scholar] [CrossRef] [PubMed]
- Heidary, M.; Nasiri, M.J.; Mirsaeidi, M.; Jazi, F.M.; Khoshnood, S.; Drancourt, M.; Darban-Sarokhalil, D. Mycobacterium avium complex infection in patients with human immunodeficiency virus: A systematic review and meta-analysis. J. Cell. Physiol. 2019, 234, 9994–10001. [Google Scholar] [CrossRef]
- Varela-Castro, L.; Barral, M.; Arnal, M.C.; Fernández de Luco, D.; Gortázar, C.; Garrido, J.M.; Sevilla, I.A. Beyond tuberculosis: Diversity and implications of non-tuberculous mycobacteria at the wildlife-livestock interface. Transbound. Emerg. Dis. 2022, 69, e2978–e2993. [Google Scholar] [CrossRef]
- Balseiro, A.; Merediz, I.; Sevilla, I.A.; García-Castro, C.; Gortázar, C.; Prieto, J.M.; Delahay, R.J. Infection of Eurasian badgers (Meles meles) with Mycobacterium avium complex (MAC) bacteria. Vet. J. 2011, 188, 231–233. [Google Scholar] [CrossRef]
- Pate, M.; Zajc, U.; Kušar, D.; Žele, D.; Vengušt, G.; Pirš, T.; Ocepek, M. Mycobacterium spp. in wild game in Slovenia. Vet. J. 2016, 208, 93–95. [Google Scholar] [CrossRef]
- Gortazar, C.; Torres, M.J.; Acevedo, P.; Aznar, J.; Negro, J.J.; de la Fuente, J.; Vicente, J. Fine-tuning the space, time, and host distribution of mycobacteria in wildlife. BMC Microbiol. 2011, 11, 27. [Google Scholar] [CrossRef]
- Matos, A.C.; Figueira, L.; Martins, M.H.; Matos, M.; Alvares, S.; Pinto, M.L.; Coelho, A.C. Disseminated Mycobacterium avium subsp. paratuberculosis infection in two wild Eurasian otters (Lutra lutra L.) from Portugal. J. Zoo Wildl. Med. 2013, 44, 193–195. [Google Scholar] [CrossRef]
- Maio, E.; Carta, T.; Balseiro, A.; Sevilla, I.A.; Romano, A.; Ortiz, J.A.; Vieira-Pinto, M.; Garrido, J.M.; de la Lastra, J.M.; Gortázar, C. Paratuberculosis in European wild rabbits from the Iberian Peninsula. Res. Vet. Sci. 2011, 91, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Rónai, Z.; Eszterbauer, E.; Csivincsik, Á.; Guti, C.F.; Dencső, L.; Jánosi, S.; Dán, Á. Detection of wide genetic diversity and several novel strains among non-avium nontuberculous mycobacteria isolated from farmed and wild animals in Hungary. J. Appl. Microbiol. 2016, 121, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Odoi, J.O.; Ohya, K.; Moribe, J.; Takashima, Y.; Sawai, K.; Taguchi, K.; Fukushi, H.; Wada, T.; Yoshida, S.; Asai, T. Isolation and antimicrobial susceptibilities of nontuberculous mycobacteria from wildlife in Japan. J. Wildl. Dis. 2020, 56, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Fischer, O.; Mátlová, L.; Bartl, J.; Dvorská, L.; Melichárek, I.; Pavlík, I. Findings of mycobacteria in insectivores and small rodents. Folia Microbiol. 2000, 45, 147–152. [Google Scholar] [CrossRef]
- Lopes, B.C.; Maciel, A.; Loiko, M.R.; Bueno, T.S.; Coppola, M.M.; Bertagnolli, A.C.; Martins, A.F.; Roehe, P.M.; Driemeier, D.; Mayer, F.Q. Molecular identification of Mycobacterium spp. isolated from Brazilian wild boars. Mol. Biol. Rep. 2021, 48, 1025–1031. [Google Scholar] [CrossRef]
- Didkowska, A.; Orłowska, B.; Krajewska-Wędzina, M.; Augustynowicz-Kopeć, E.; Brzezińska, S.; Żygowska, M.; Wiśniewski, J.; Kaczor, S.; Welz, M.; Olech, W.; et al. Microbiological and molecular monitoring for bovine tuberculosis in the Polish population of European bison (Bison bonasus). Ann. Agric. Environ. Med. 2021, 28, 575–578. [Google Scholar] [CrossRef]
- Radulski, Ł.; Krajewska-Wędzina, M.; Lipiec, M.; Szulowski, K. Infection of a Free-Living Wild Boar (Sus scrofa) with a Bacterium from the Mycobacterium kansasii Complex. Animals 2022, 12, 964. [Google Scholar] [CrossRef]
- Majchrzak, M.; Kaczmarkowska, A.; Didkowska, A.; Brzezińska, S.; Orłowska, B.; Klich, D.; Augustynowicz-Kopeć, E.; Anusz, K.; Parniewski, P. MIRU-VNTR Typing of Atypical Mycobacteria Isolated from the Lymph Nodes of Slaughtered Pigs from Poland. Pathogens 2022, 11, 495. [Google Scholar] [CrossRef]
- Imperiale, B.; Moyano, R.; Di Giulio, A.; Romero, M.; Pinedo, A.; Santangelo, M.P.; Travería, G.E.; Morcillo, N.S.; Romano, M.I. Genetic diversity of Mycobacterium avium complex strains isolated in Argentina by MIRU-VNTR. Epidemiol. Infect. 2017, 145, 1382–1391. [Google Scholar] [CrossRef]
- Thibault, V.C.; Grayon, M.; Boschiroli, M.L.; Hubbans, C.; Overduin, P.; Stevenson, K.; Gutierrez, M.C.; Supply, P.; Biet, F. New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: Comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J. Clin. Microbiol. 2007, 45, 2404–2410. [Google Scholar] [CrossRef]
- VerCauteren, K.C.; Atwood, T.C.; DeLiberto, T.J.; Smith, H.J.; Stevenson, J.S.; Thomsen, B.V.; Gidlewski, T.; Payeur, J. Surveillance of coyotes to detect bovine tuberculosis, Michigan. Emerg. Infect. Dis. 2008, 14, 1862–1869. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, M.; Lipiec, M.; Zabost, A.; Augustynowicz-Kopeć, E.; Szulowski, K. Bovine tuberculosis in a wild boar (Sus scrofa) in Poland. J. Wildl. Dis. 2014, 50, 1001–1002. [Google Scholar] [CrossRef] [PubMed]
- Orłowska, B.; Augustynowicz-Kopeć, E.; Krajewska, M.; Zabost, A.; Welz, M.; Kaczor, S.; Anusz, K. Mycobacterium caprae transmission to free-living grey wolves (Canis lupus) in the Bieszczady Mountains in Southern Poland. Eur. J. Wildl. Res. 2017, 63, 21. [Google Scholar] [CrossRef]
- Orłowska, B.; Krajewska-Wędzina, M.; Augustynowicz-Kopeć, E.; Kozińska, M.; Brzezińska, S.; Zabost, A.; Didkowska, A.; Welz, M.; Kaczor, S.; Żmuda, P.; et al. Epidemiological characterization of Mycobacterium caprae strains isolated from wildlife in the Bieszczady Mountains, on the border of Southeast Poland. BMC Vet. Res. 2020, 16, 362. [Google Scholar] [CrossRef] [PubMed]
- Lipiec, M. Bovine tuberculosis, diagnosis, control, current status, comments. Monogr. Natl. Vet. Res. Inst. 2016, 1, 7–121. (In Polish) [Google Scholar]
- Cochard, T.; Branger, M.; Supply, P.; Sreevatsan, S.; Biet, F. MAC-INMV-SSR: A web application dedicated to genotyping members of Mycobacterium avium complex (MAC) including Mycobacterium avium subsp. paratuberculosis strains. Infect. Genet. Evol. 2020, 77, 104075. [Google Scholar] [CrossRef]
- Dhama, K.; Mahendran, M.; Tiwari, R.; Dayal Singh, S.; Kumar, D.; Singh, S.; Sawant, P.M. Tuberculosis in Birds: Insights into the Mycobacterium avium Infections. Vet. Med. Int. 2011, 2011, 712369. [Google Scholar] [CrossRef]
- Ledwoń, A.; Napiórkowska, A.; Augustynowicz-Kopeć, E.; Szeleszczuk, P. Drug Susceptibility of Non-tuberculous Strains of Mycobacterium Isolated from Birds from Poland. Pol. J. Microbiol. 2018, 67, 487–492. [Google Scholar] [CrossRef]
- Richomme, C.; Lesellier, S.; Salguero, F.J.; Barrat, J.L.; Boucher, J.-M.; Reyes-Reyes, J.D.; Hénault, S.; De Cruz, K.; Tambosco, J.; Michelet, L.; et al. Experimental Infection of Captive Red Foxes (Vulpes vulpes) with Mycobacterium bovis. Microorganisms 2022, 10, 380. [Google Scholar] [CrossRef]
- Bruning-Fann, C.S.; Schmitt, S.M.; Fitzgerald, S.D.; Payeur, J.B.; Whipple, D.L.; Cooley, T.M.; Carlson, T.; Friedrich, P. Mycobacterium bovis in coyotes in Michigan. J. Wildl. Dis. 1998, 34, 632–636. [Google Scholar] [CrossRef]
- Delahay, R.J.; De Leeuw, A.N.; Barlow, A.M.; Clifton-hadley, R.S.; Cheeseman, C.L. The status of Mycobacterium bovis infection in UK wild mammals: A review. Vet. J. 2002, 164, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Delahay, R.J.; Smith, G.C.; Barlow, A.M.; Walker, N.; Harris, A.; Clifton-Hadley, R.S.; Cheeseman, C.L. Bovine tuberculosis infection in wild mammals in the South-West region of England: A survey of prevalence and a semi-quantitative assessment of the relative risks to cattle. Vet. J. 2007, 173, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Berentsen, A.R.; Dunbar, M.R.; Johnson, S.R.; Robbe-Austerman, S.; Martinez, L.; Jones, R.L. Active use of coyotes (Canis latrans) to detect Bovine Tuberculosis in northeastern Michigan, USA. Vet. Microbiol. 2011, 151, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Romero, B.; Aranaz, A.; Sandoval, A.; Alvarez, A.; de Juan, L.; Bezos, J.; Sanchez, C.; Galka, M.; Fernandez, P.; Mateos, A.; et al. Persistence and molecular evolution of Mycobacterium bovis population from cattle and wildlife in Donana National Park revealed by genotype variation. Vet. Microbiol. 2008, 132, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Garrido, J.M.; Vicente, J.; Carrasco-Garcia, R.; Galindo, R.C.; Minguijon, E.; Ballesteros, C.; Aranaz, A.; Romero, B.; Sevilla, I.; Juste, R.; et al. Experimental infection of Eurasian wild boar with Mycobacterium avium subsp. Avium. Vet. Microbiol. 2010, 144, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Tschopp, R.; Berg, S.; Argaw, K.; Gadisa, E.; Habtamu, M.; Schelling, E.; Young, D.; Aseffa, A.; Zinsstag, J. Bovine tuberculosis in Ethiopian wildlife. J. Wildl. Dis. 2010, 46, 753–762. [Google Scholar] [CrossRef]
- Shitaye, J.E.; Matlova, L.; Horvathova, A.; Moravkova, M.; Dvorska-Bartosova, L.; Treml, F.; Lamka, J.; Pavlik, I. Mycobacterium avium subsp. avium distribution studied in a naturally infected hen flock and in the environment by culture, serotyping and IS901 RFLP methods. Vet. Microbiol. 2008, 127, 155–164. [Google Scholar] [CrossRef]
- Ikonomopoulos, J.; Fragkiadaki, E.; Liandris, E.; Sotirakoglou, K.; Xylouri, E.; Gazouli, M. Estimation of the spread of pathogenic mycobacteria in organic broiler farms by the polymerase chain reaction. Vet. Microbiol. 2009, 133, 278–282. [Google Scholar] [CrossRef][Green Version]
- Dvorska, L.; Matlova, L.; Bartos, M.; Parmova, I.; Bartl, J.; Svastova, P.; Bull, T.J.; Pavlik, I. Study of Mycobacterium avium complex strains isolated from cattle in the Czech Republic between 1996 and 2000. Vet. Microbiol. 2004, 99, 239–250. [Google Scholar] [CrossRef]
- Chiers, K.; Deschaght, P.; De Baere, T.; Dabrowski, S.; Kotlowski, R.; De Clercq, D.; Ducatelle, R.; Vaneechoutte, M. Isolation and identification of Mycobacterium avium subspecies silvaticum from a horse. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 303–307. [Google Scholar] [CrossRef]
- Schrenzel, M.; Nicolas, M.; Witte, C.; Papendick, R.; Tucker, T.; Keener, L.; Sutherland-Smith, M.; Lamberski, N.; Orndorff, D.; Heckard, D.; et al. Molecular epidemiology of Mycobacterium avium subsp. avium and Mycobacterium intracellulare in captive birds. Vet. Microbiol. 2008, 126, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Durnez, L.; Suykerbuyk, P.; Nicolas, V.; Barrière, P.; Verheyen, E.; Johnson, C.R.; Leirs, H.; Portaels, F. Terrestrial Small Mammals as Reservoirs of Mycobacterium ulcerans in Benin. Appl. Environ. Microbiol. 2010, 76, 4574–4577. [Google Scholar] [CrossRef] [PubMed]
- Durnez, L.; Katakweba, A.; Sadiki, H.; Katholi, C.R.; Kazwala, R.R.; Machang’u, R.R.; Portaels, F.; Leirs, H. Mycobacteria in terrestrial small mammals on cattle farms in Tanzania. Vet. Med. Int. 2011, 2011, 495074. [Google Scholar] [CrossRef] [PubMed]
- Eaton, T.; Falkinham, J.O.; Aisu, T.O.; Daniel, T.M. Isolation and characteristics of Mycobacterium avium complex from water and soil samples in Uganda. Tuber. Lung Dis. 1995, 76, 570–574. [Google Scholar] [CrossRef]
- Falkinham, J.O., 3rd. Surrounded by mycobacteria: Nontuberculous mycobacteria in the human environment. J. Appl. Microbiol. 2009, 107, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Fischer, O.A.; Matlova, L.; Bartl, J.; Dvorska, L.; Svastova, P.; du Maine, R.; Melicharek, I.; Bartos, M.; Pavlik, I. Earthworms (Oligochaeta: Lumbricidae) and mycobacteria. Vet. Microbiol. 2003, 91, 325–338. [Google Scholar] [CrossRef]
- Klanicova-Zalewska, B.; Slana, I. Presence and persistence of Mycobacterium avium and other nontuberculous mycobacteria in animal tissues and derived foods: A review. Meat Sci. 2014, 98, 835–841. [Google Scholar] [CrossRef]
- Orłowska, B.; Anusz, K.; Krajewska, M.; Augustynowicz-Kopeć, E.; Zabost, A.; Nowicki, M. Recognition of the Mycobacterium tuberculosis complex reservoirs among free-ranging red deer (Cervus elaphus) in the Bieszczady region (south-eastern Poland). In Proceedings of the XIV Middle European Buiatrics Congress, Warsaw, Poland, 25–27 May 2014; p. 167. [Google Scholar]
- Welz, M. The Epizootic Situation among Livestock and the Wildlife in the Area of Bieszczady Mountains Considering the Mycobacterium bovis Infections. PhD Thesis, University of Life Sciences, Warsaw, Poland, 2010. [Google Scholar]
- Shin, M.K.; Shin, S.J. Genetic Involvement of Mycobacterium avium Complex in the Regulation and Manipulation of Innate Immune Functions of Host Cells. Int. J. Mol. Sci. 2021, 22, 3011. [Google Scholar] [CrossRef]
- Brown-Elliott, B.A.; Nash, K.A.; Wallace, R.J. Antimicrobial Susceptibility Testing, Drug Resistance Mechanisms, and Therapy of Infections with Nontuberculous Mycobacteria. Clin. Microbiol. Rev. 2012, 25, 545–582. [Google Scholar] [CrossRef]
- Couto, C.; Rossetti, S.; Schlaen, A.; Hurtado, E.; D’Alessandro, L.; Goldstein, D.A. Chronic postoperative Mycobacterium gordonae endophthalmitis in a patient with phakic intraocular lens. Ocul. Immunol. Inflamm. 2013, 21, 491–494. [Google Scholar] [CrossRef]
- Yazbek, S.; Aggarwal, A.; Simpson, D.M.; Som, P.M. Imaging findings of atypical mycobacterial infection in the temporal bone. Clin. Imaging 2013, 37, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Chien, J.Y.; Lai, C.C.; Sheng, W.H.; Yu, C.J.; Hsueh, P.R. Pulmonary Infection and Colonization with Nontuberculous Mycobacteria, Taiwan, 2000–2012. Emerg. Infect. Dis. 2014, 20, 1382–1385. [Google Scholar] [CrossRef] [PubMed]
- Mirsaeidi, M.; Machado, R.F.; Garcia, J.G.N.; Schraufnagel, D.E. Nontuberculous Mycobacterial Disease Mortality in the United States, 1999–2010: A Population-Based Comparative Study. PLoS ONE 2014, 9, e91879. [Google Scholar] [CrossRef] [PubMed]
- Queiros, J.; Alvarez, J.; Carta, T.; Mateos, A.; Ortiz, J.A.; Fernández-de-Mera, I.G.; Martín-Hernando, M.P.; Gortázar, C. Unexpected high responses to tuberculin skin-test in farmed red deer: Implications for tuberculosis control. Prev. Vet. Med. 2012, 104, 327–334. [Google Scholar] [CrossRef]
- Norby, B.; Fosgate, G.T.; Manning, E.J.B.; Collins, M.T.; Roussel, A.J. Environmental mycobacteria in soil and water on beef ranches: Association between presence of cultivable mycobacteria and soil and water physicochemical characteristics. Vet. Microbiol. 2007, 124, 153–159. [Google Scholar] [CrossRef]
| No. (Isolate Number) | The Origin of the Strain | Number of Copies of MIRU-VNTR Region | Subspecies Assignment | IS901 | IS900 | IS1245 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TR 292 | TR x3 | TR 25 | TR 47 | TR 3 | TR 7 | TR 10 | TR 32 | ||||||
| 1 (169) | Beech marten | 1 | 2 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 2 (23 b) | European badger | 1 | 3 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 3 (W17) | Grey wolf | 2 | 2 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 4 (603) | Red fox | 2 | 3 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 5 (493) | Red fox | 2 | 3 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 6 (494) | Red fox | 2 | 3 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 7 (522) | Red fox | 2 | 4 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 8 (56) | Red fox | 2 | 4 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 9 (108) | Red fox | 2 | 4 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 10 (579) | Red fox | 2 | 4 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 11 (615) | Red fox | 2 | 4 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 12 (619) | Red fox | 2 | 4 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 13 (582) | Red fox | 2 | 4 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 14 (517) | Red fox | 2 | 4 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 15 (593) | Red fox | 2 | 4 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 16 (523) | Red fox | 2 | 4 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 17 (629) | Red fox | 2 | 4 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 18 (488) | Red fox | 2 | 4 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 19 (513) | Red fox | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 20 (510) | Red fox | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 21 (509) | Red fox | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 22 (498) | Red fox | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 23 (512) | Red fox | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 24 (461) | Red fox | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 25 (130) | Red fox | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 26 (1.4.5) | European badger | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 27 (529) | Red fox | 2 | 5 | 1 | 3 | 1 | 2 | 3 | 7 | MAA | + | - | + |
| 28 (1 W) | Grey wolf | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 29 (171) | Red fox | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 30 (218) | Red fox | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 31 (Pt3) | Buzzard | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 32 (178) | Red fox | 2 | 5 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 33 (W20) | Grey wolf | 3 | 3 | 1 | 3 | 1 | 2 | 2 | 7 | MAA | + | - | + |
| 34 (520) | Red fox | - | - | - | - | - | - | - | - | NA | - | - | - |
| 35 (83.4) | Red fox | - | - | - | - | - | - | - | - | NA | - | - | - |
| 36 (447) | Red fox | - | - | - | - | - | - | - | - | NA | - | - | - |
| 37 (235) | Red fox | - | - | - | - | - | - | - | - | NA | - | - | - |
| 38 (W32) | Grey wolf | 2 | 5 | 3 | 2 | 1 | 2 | 2 | 8 | MAH | - | - | + |
| 39 (W18) | Grey wolf | 2 | 2 | 1 | 3 | 1 | 2 | - | 7 | NA | + | - | - |
| 40 (134) | European badger | 2 | 5 | 1 | 3 | 1 | 2 | - | - | NA | + | - | - |
| 41 (W15) | Grey wolf | - | - | 1 | 3 | 1 | 2 | - | 7 | NA | + | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orłowska, B.; Majchrzak, M.; Didkowska, A.; Anusz, K.; Krajewska-Wędzina, M.; Zabost, A.; Brzezińska, S.; Kozińska, M.; Augustynowicz-Kopeć, E.; Urbańska, K.; et al. Mycobacterial Interspersed Repeat Unit–Variable Number Tandem Repeat Typing of Mycobacterium avium Strains Isolated from the Lymph Nodes of Free-Living Carnivorous Animals in Poland. Pathogens 2023, 12, 1184. https://doi.org/10.3390/pathogens12091184
Orłowska B, Majchrzak M, Didkowska A, Anusz K, Krajewska-Wędzina M, Zabost A, Brzezińska S, Kozińska M, Augustynowicz-Kopeć E, Urbańska K, et al. Mycobacterial Interspersed Repeat Unit–Variable Number Tandem Repeat Typing of Mycobacterium avium Strains Isolated from the Lymph Nodes of Free-Living Carnivorous Animals in Poland. Pathogens. 2023; 12(9):1184. https://doi.org/10.3390/pathogens12091184
Chicago/Turabian StyleOrłowska, Blanka, Marta Majchrzak, Anna Didkowska, Krzysztof Anusz, Monika Krajewska-Wędzina, Anna Zabost, Sywia Brzezińska, Monika Kozińska, Ewa Augustynowicz-Kopeć, Kaja Urbańska, and et al. 2023. "Mycobacterial Interspersed Repeat Unit–Variable Number Tandem Repeat Typing of Mycobacterium avium Strains Isolated from the Lymph Nodes of Free-Living Carnivorous Animals in Poland" Pathogens 12, no. 9: 1184. https://doi.org/10.3390/pathogens12091184
APA StyleOrłowska, B., Majchrzak, M., Didkowska, A., Anusz, K., Krajewska-Wędzina, M., Zabost, A., Brzezińska, S., Kozińska, M., Augustynowicz-Kopeć, E., Urbańska, K., Welz, M., & Parniewski, P. (2023). Mycobacterial Interspersed Repeat Unit–Variable Number Tandem Repeat Typing of Mycobacterium avium Strains Isolated from the Lymph Nodes of Free-Living Carnivorous Animals in Poland. Pathogens, 12(9), 1184. https://doi.org/10.3390/pathogens12091184








