Assembly, Annotation, and Comparative Whole Genome Sequence of Fusarium verticillioides Isolated from Stored Maize Grains
Abstract
:1. Introduction
2. Results
2.1. Genome Sequence, Assembly, Statistics, and Annotation
2.2. Identification of Repetitive Elements (REs) and Simple Sequence Repeats (SSRs)
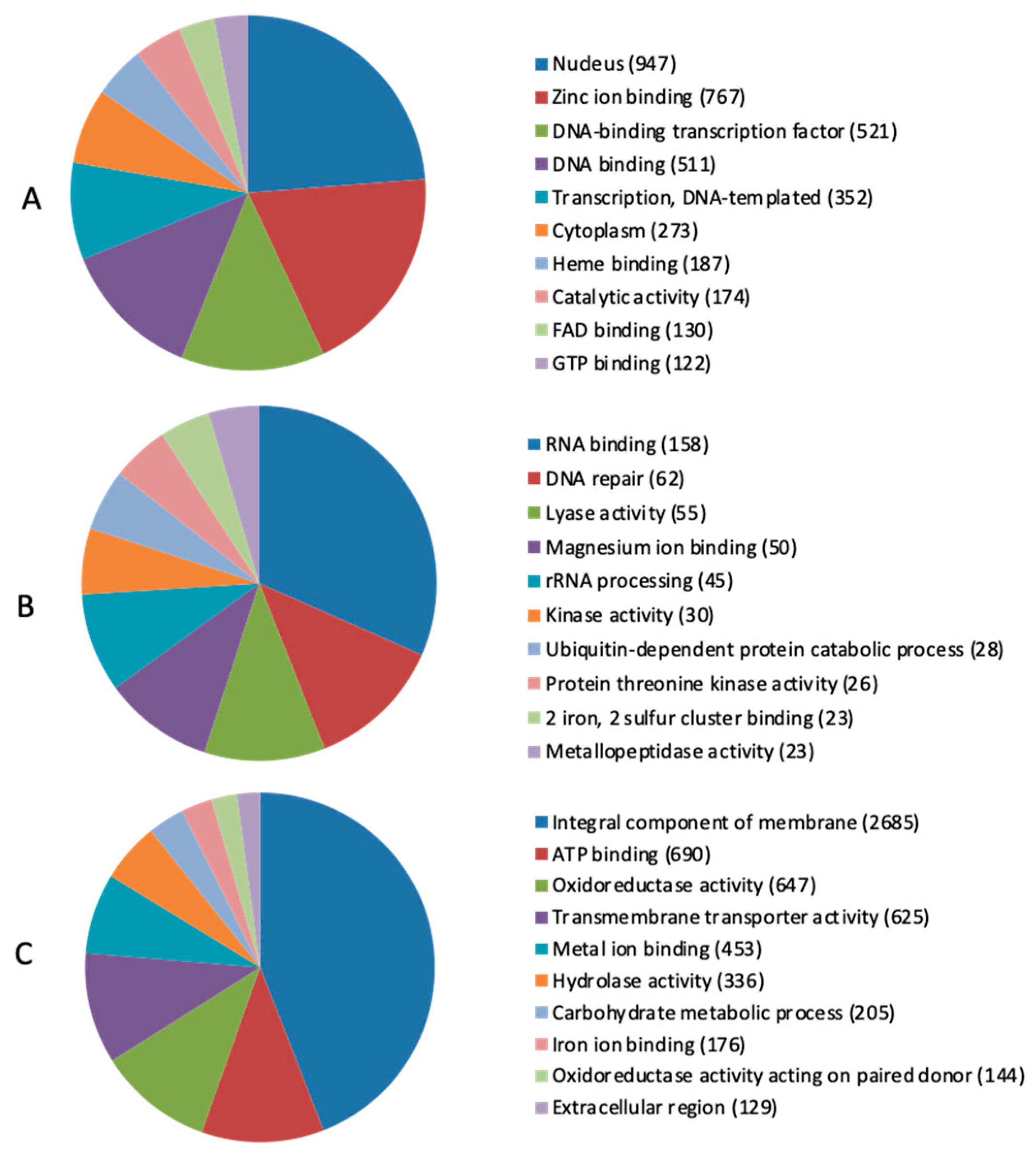
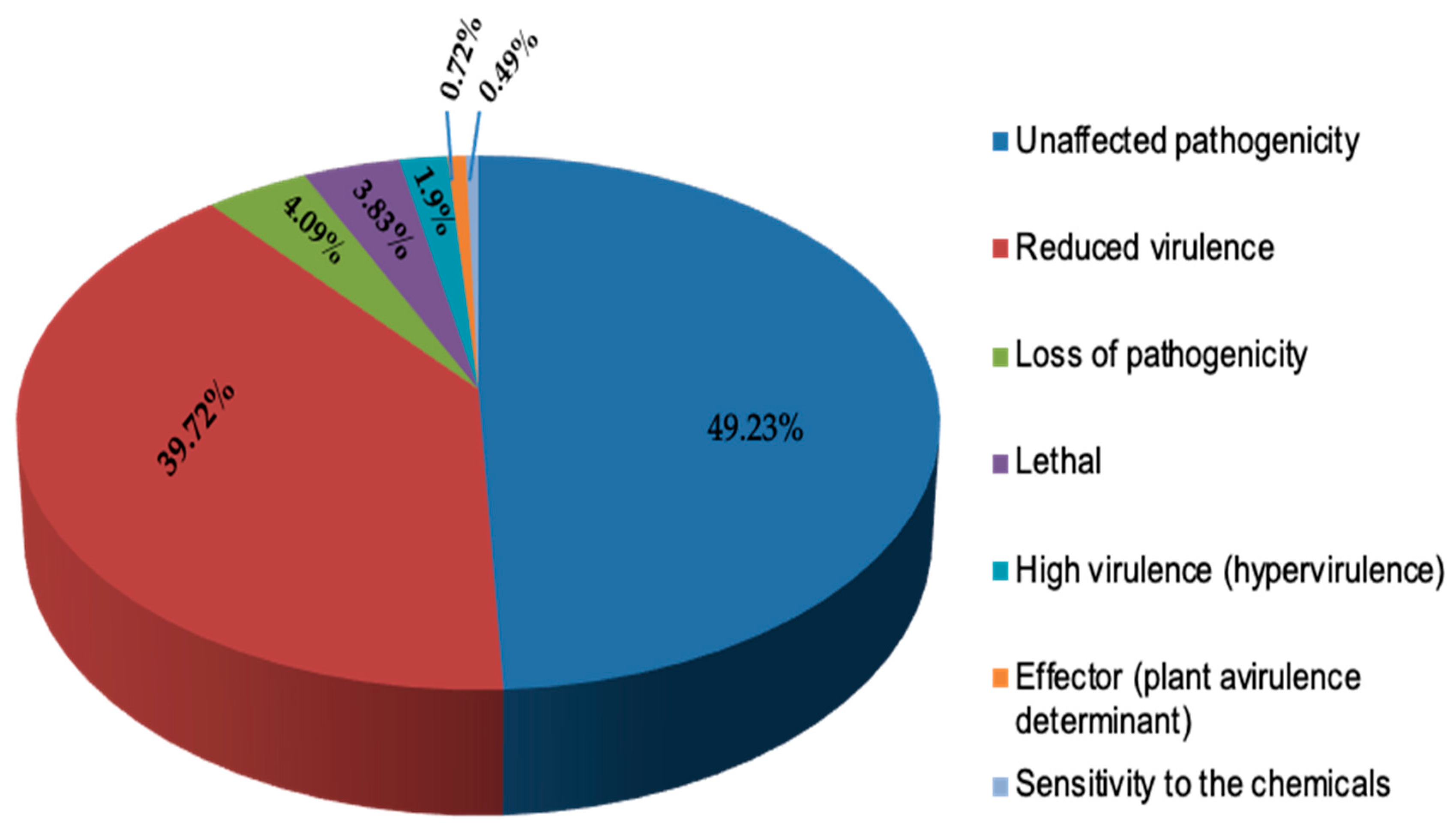
2.3. Secretome Prediction and Host–Pathogen Interaction Analysis
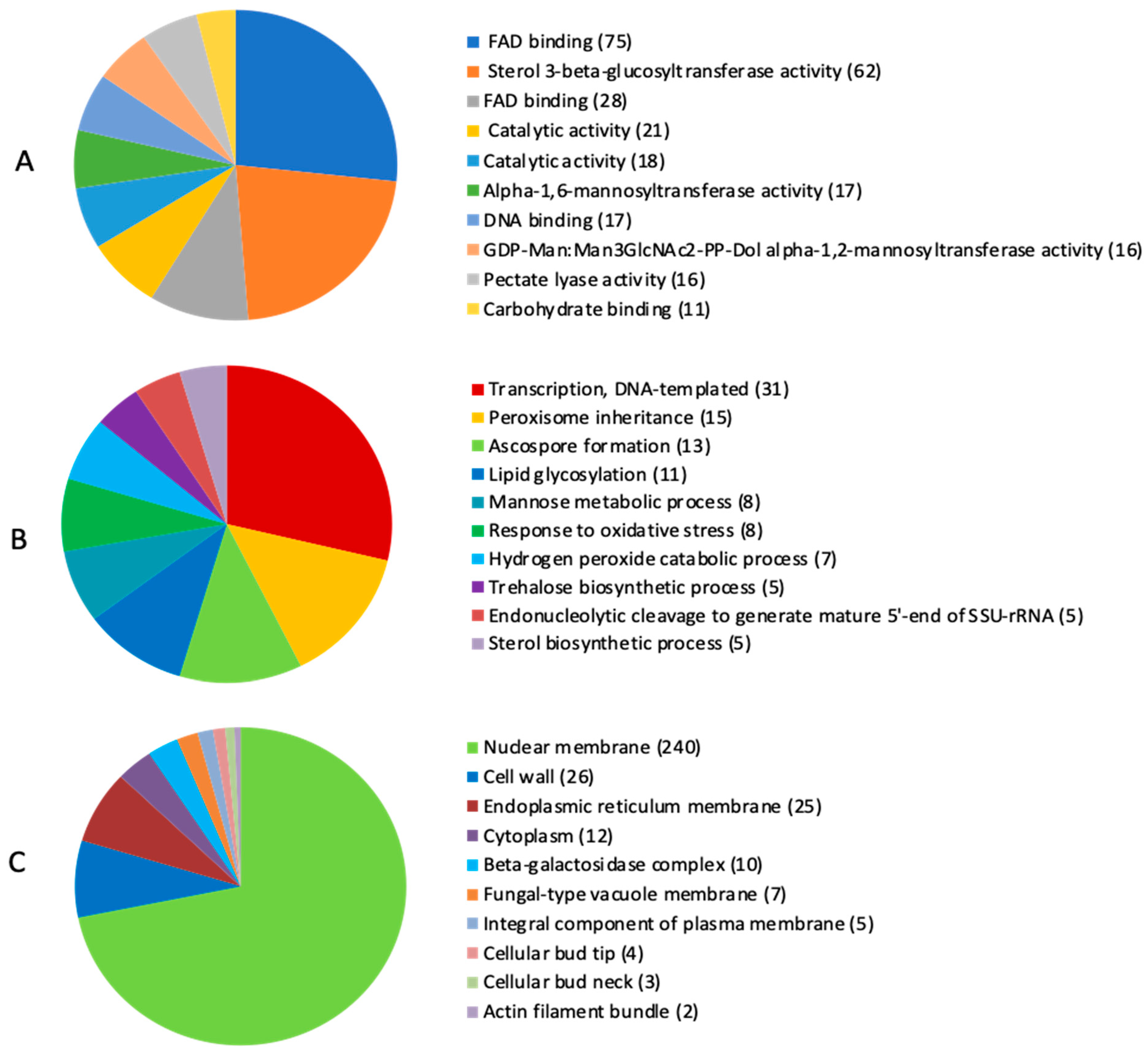
2.4. Identification of Carbohydrate-Active Enzymes (CAZymes) and Mycotoxin Biosynthetic Genes
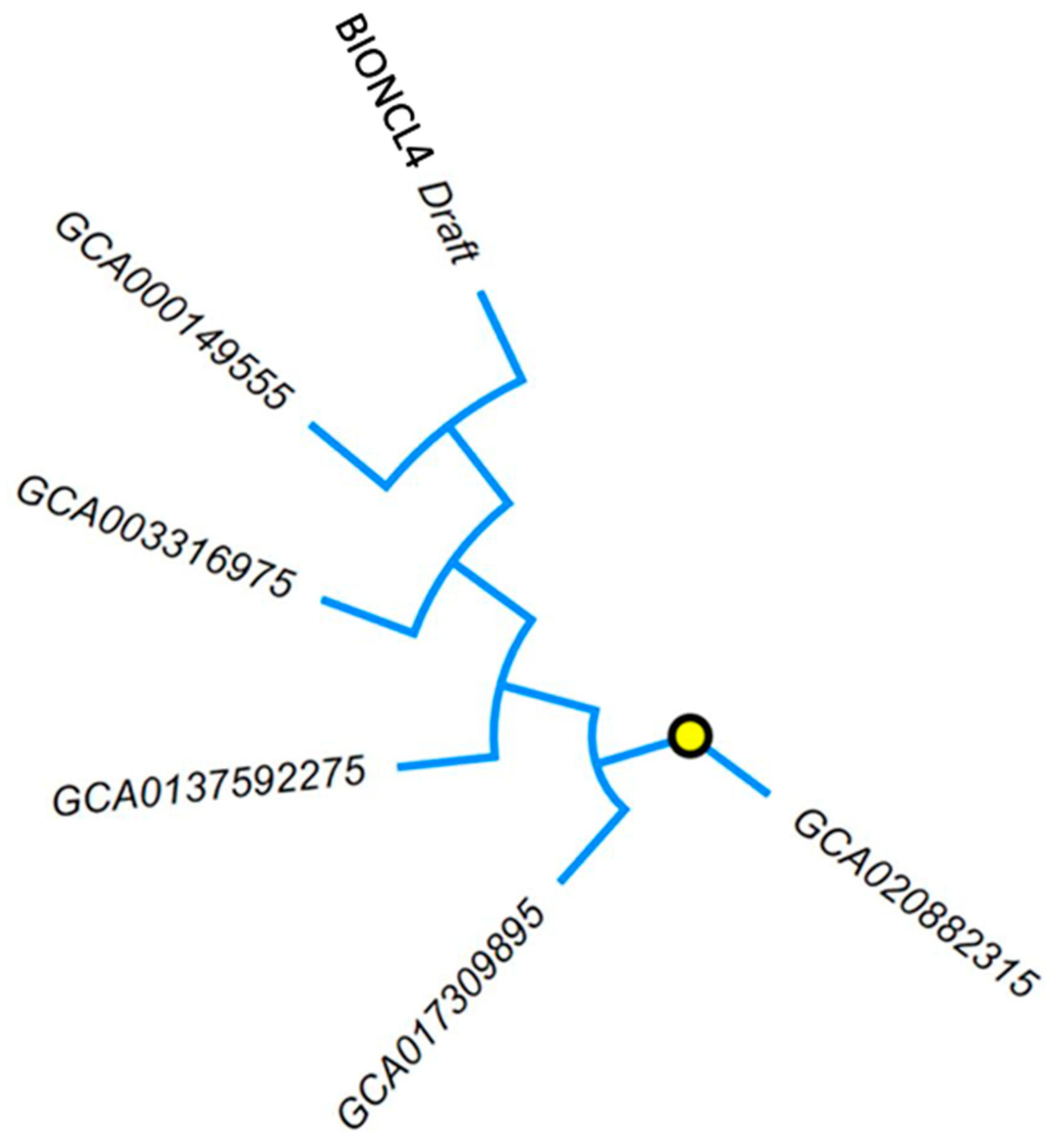
2.5. Comparative Draft Genome Analysis of BIONCL4 Strain
3. Discussion
4. Materials and Methods
4.1. Culture Conditions and DNA Isolation
4.2. Genome Sequencing and Assembly
4.3. Gene Prediction and Annotation
4.4. Identification of Repetitive Elements (REs) and Single Nucleotide Polymorphism (SNP)
4.5. Secretome Prediction and Functional Genomic Analysis
4.6. Analysis of Orthologous Gene Families and Mycotoxin Biosynthetic Gene Identification
4.7. Comparative Phylogenetic Analysis of Fusarium Genomes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, Q.; Balint-Kurti, P.; Xu, M. Quantitative Disease Resistance: Dissection and Adoption in Maize. Mol. Plant 2017, 10, 402–413. [Google Scholar] [CrossRef] [Green Version]
- Qiu, J.; Lu, Y.; He, D.; Lee, Y.W.; Ji, F.; Xu, J.; Shi, J. Fusarium Fujikuroi Species Complex Associated with Rice, Maize, and Soybean From Jiangsu Province, China: Phylogenetic, Pathogenic, and Toxigenic Analysis. Plant Dis. 2020, 104, 2193–2201. [Google Scholar] [CrossRef]
- Dastjerdi, R.; Karlovsky, P. Systemie Infection of Maize, Sorghum, Rice, and Beet Seedlings with Fumonisin-Producing and Nonproducing Fusarium verticillioides Strains. Plant Pathol. J. 2015, 31, 334–342. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.; Son, J.H.; Park, J.C.; Kim, K.H.; Yoon, Y.M.; Cheong, Y.K.; Kim, K.H.; Hyun, J.N.; Park, C.S.; Dill-Macky, R.; et al. Comparative Pathogenicity of Fusarium graminearum Isolates from Wheat Kernels in Korea. Plant Pathol. J. 2018, 34, 347. [Google Scholar] [CrossRef]
- Ekhuemelo, C.; Igbor, H.U.; Ocheje, S.J. Screening of Cowpea (Vigna Unguiculata (L.) Walp) Varieties for Resistance to Leaf Spot in Southern Guinea Savannah Agro- Ecology of Nigeria. Niger. J. Biotechnol. 2020, 36, 9–20. [Google Scholar] [CrossRef]
- Puértolas, A.; Boa, E.; Bonants, P.J.M.; Woodward, S. Survival of Phytophthora cinnamomi and Fusarium verticillioides in Commercial Potting Substrates for Ornamental Plants. J. Phytopathol. 2018, 166, 484–493. [Google Scholar] [CrossRef]
- Aiyaz, M.; Thimmappa Divakara, S.; Mudili, V.; George Moore, G.; Kumar Gupta, V.; Yli-Mattila, T.; Chandra Nayaka, S.; Ramachandrappa Niranjana, S. Molecular Diversity of Seed-Borne Fusarium Species Associated with Maize in India. Curr. Genomics 2015, 17, 132–144. [Google Scholar] [CrossRef] [Green Version]
- Deepa, N.; Nagaraja, H.; Sreenivasa, M.Y. Prevalence of Fumonisin Producing Fusarium verticillioides Associated with Cereals Grown in Karnataka (India). Food Sci. Hum. Wellness 2016, 5, 156–162. [Google Scholar] [CrossRef] [Green Version]
- Nagaraja, H.; Chennappa, G.; Poorna Chandra Rao, K.; Mahadev Prasad, G.; Sreenivasa, M.Y. Diversity of Toxic and Phytopathogenic Fusarium Species Occurring on Cereals Grown in Karnataka State, India. 3 Biotech 2016, 6, 57. [Google Scholar] [CrossRef] [Green Version]
- Bashyal, B.M.; Rawat, K.; Sharma, S.; Kulshreshtha, D.; Gopala Krishnan, S.; Singh, A.K.; Dubey, H.; Solanke, A.U.; Sharma, T.R.; Aggarwal, R. Whole Genome Sequencing of Fusarium Fujikuroi Provides Insight into the Role of Secretory Proteins and Cell Wall Degrading Enzymes in Causing Bakanae Disease of Rice. Front. Plant Sci. 2017, 8, 2013. [Google Scholar] [CrossRef] [Green Version]
- Navale, V.; Penugonda, S.; Vamkudoth, K.R. Prevalence of zearalenone producing Fusarium species associated with finger millet. Indian Phytopathol. 2022, 75, 367–375. [Google Scholar] [CrossRef]
- Divakara, S.T.; Santosh, P.; Aiyaz, M.; Ramana, M.V.; Hariprasad, P.; Nayaka, S.C.; Niranjana, S.R. Molecular identification and characterization of Fusarium spp. associated with sorghum seeds. J. Sci. Food Agric. 2014, 94, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Gupta, V.K.; Misra, A.K.; Gaur, R.; Bajpai, V.; Issar, S. Current Status of Fusarium Infection in Human and Animal. Asian, J. Anim. Vet. Adv. 2011, 6, 201–227. [Google Scholar] [CrossRef] [Green Version]
- Walther, G.; Stasch, S.; Kaerger, K.; Hamprecht, A.; Roth, M.; Cornely, O.A.; Geerling, G.; Mackenzie, C.R.; Kurzai, O.; Von Lilienfeld-Toal, M. Fusarium Keratitis in Germany. J. Clin. Microbiol. 2017, 55, 2983. [Google Scholar] [CrossRef] [Green Version]
- Tupaki-Sreepurna, A.; Thanneru, V.; Natarajan, S.; Sharma, S.; Gopi, A.; Sundaram, M.; Kindo, A.J. Phylogenetic Diversity and In Vitro Susceptibility Profiles of Human Pathogenic Members of the Fusarium fujikuroi Species Complex Isolated from South India. Mycopathologia 2018, 183, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Leyva-Madrigal, K.Y.; Sandoval-Castro, E.; Calderón-Vázquez, C.L.; Larralde-Corona, C.P.; Maldonado-Mendoza, I.E. Pathogenic and Genetic Variability of Fusarium verticillioides from Maize in Northern Mexico. Can. J. Plant Pathol. 2017, 39, 486–496. [Google Scholar] [CrossRef]
- Navale, V.; Vamkudoth, K.R.; Ajmera, S.; Dhuri, V. Aspergillus Derived Mycotoxins in Food and the Environment: Prevalence, Detection, and Toxicity. Toxicol. Rep. 2021, 8, 1008–1030. [Google Scholar] [CrossRef]
- Sharma, G.; Kumar Pinnaka, A.; Damodara Shenoy, B. Infra-Specific Diversity of Colletotrichum truncatum Associated with Chilli Anthracnose in India Based on Microsatellite Marker Analysis. Arch. Phytopathol. Plant Prot. 2014, 47, 2509–2523. [Google Scholar] [CrossRef]
- Pfordt, A.; Schiwek, S.; Rathgeb, A.; Rodemann, C.; Bollmann, N.; Buchholz, M.; Karlovsky, P.; von Tiedemann, A. Occurrence, Pathogenicity, and Mycotoxin Production of Fusarium temperatum in Relation to Other Fusarium Species on Maize in Germany. Pathogens 2020, 9, 864. [Google Scholar] [CrossRef]
- Ji, F.; He, D.; Olaniran, A.O.; Mokoena, M.P.; Xu, J.; Shi, J. Occurrence, Toxicity, Production and Detection of Fusarium Mycotoxin: A Review. Food Prod. Process. Nutr. 2019, 1, 6. [Google Scholar] [CrossRef]
- Silva, J.J.; Viaro, H.P.; Ferranti, L.S.; Oliveira, A.L.M.; Ferreira, J.M.; Ruas, C.F.; Ono, E.Y.S.; Fungaro, M.H.P. Genetic Structure of Fusarium verticillioides Populations and Occurrence of Fumonisins in Maize Grown in Southern Brazil. Crop Prot. 2017, 99, 160–167. [Google Scholar] [CrossRef]
- Kamle, M.; Mahato, D.K.; Devi, S.; Lee, K.E.; Kang, S.G.; Kumar, P. Fumonisins: Impact on Agriculture, Food, and Human Health and Their Management Strategies. Toxins 2019, 11, 328. [Google Scholar] [CrossRef] [Green Version]
- Ferrigo, D.; Raiola, A.; Causin, R. Fusarium Toxins in Cereals: Occurrence, Legislation, Factors Promoting the Appearance and Their Management. Molecules 2016, 21, 627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Wei, Z.; Wang, Y.; Long, M.; Wu, W.; Kuca, K. Fumonisin B1: Mechanisms of Toxicity and Biological Detoxification Progress in Animals. Food Chem. Toxicol. 2021, 149, 111977. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer. Some Traditional Herbal Medicines, Some Mycotoxins, Naphthalene and Styrene; This Publication Represents the Views and Expert Opinions of an IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, Which Met in Lyon, 12–19 February 2002; IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; IARC: Lyon, France, 2002; ISBN 978-92-832-1282-9. [Google Scholar]
- Sun, G.; Wang, S.; Hu, X.; Su, J.; Huang, T.; Yu, J.; Tang, L.; Gao, W.; Wang, J.S. Fumonisin B1 Contamination of Home-Grown Corn in High-Risk Areas for Esophageal and Liver Cancer in China. Food Addit. Contam. 2007, 24, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Szabó, A.; Szabó-Fodor, J.; Kachlek, M.; Mézes, M.; Balogh, K.; Glávits, R.; Ali, O.; Zeebone, Y.Y.; Kovács, M. Dose and Exposure Time-Dependent Renal and Hepatic Effects of Intraperitoneally Administered Fumonisin B 1 in Rats. Toxins 2018, 10, 465. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Sarkar, P.; Chattopadhyay, A. Metabolic Depletion of Sphingolipids Reduces Cell Surface Population of the Human Serotonin1AReceptor Due to Impaired Trafficking. ACS Chem. Neurosci. 2021, 12, 1189–1196. [Google Scholar] [CrossRef]
- Fanning, S.; Proos, S.; Jordan, K.; Srikumar, S. A Review on the Applications of Next Generation Sequencing Technologies as Applied to Food-Related Microbiome Studies. Front. Microbiol. 2017, 8, 1829. [Google Scholar] [CrossRef]
- King, R.; Urban, M.; Hammond-Kosack, M.C.U.; Hassani-Pak, K.; Hammond-Kosack, K.E. The Completed Genome Sequence of the Pathogenic Ascomycete Fungus Fusarium graminearum. BMC Genomics 2015, 16, 544. [Google Scholar] [CrossRef] [Green Version]
- Ravalason, H.; Grisel, S.; Chevret, D.; Favel, A.; Berrin, J.G.; Sigoillot, J.C.; Herpoël-Gimbert, I. Fusarium verticillioides Secretome as a Source of Auxiliary Enzymes to Enhance Saccharification of Wheat Straw. Bioresour. Technol. 2012, 114, 589–596. [Google Scholar] [CrossRef]
- Lanubile, A.; Ferrarini, A.; Maschietto, V.; Delledonne, M.; Marocco, A.; Bellin, D. Functional Genomic Analysis of Constitutive and Inducible Defense Responses to Fusarium verticillioides Infection in Maize Genotypes with Contrasting Ear Rot Resistance. BMC Genomics 2014, 15, 710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardiner, D.M. Genome Sequences of Three Isolates of Fusarium verticillioides. Microbiol. Resour. Announc. 2018, 7, e00918-18. [Google Scholar] [CrossRef] [Green Version]
- Raman, T.; Raj, E.E.; Muthukathan, G.; Loganathan, M.; Periyasamy, P.; Natesh, M.; Manivasakan, P.; Kotteeswaran, S.; Rajendran, S.; Subbaraya, U. Comparative Whole-Genome Sequence Analyses of Fusarium Wilt Pathogen (Foc R1, STR4 and TR4) Infecting Cavendish (AAA) Bananas in India, with a Special Emphasis on Pathogenicity Mechanisms. J. Fungi 2021, 7, 717. [Google Scholar] [CrossRef] [PubMed]
- Thangavelu, R.; Raj, E.E.; Pushpakanth, P.; Loganathan, M.; Uma, S. Draft Genome of Fusarium oxysporum f. Sp. cubense Strain Tropical Race-4 Infecting Cavendish (AAA) Group of Banana in India. Plant Dis. 2021, 105, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 Fungal Pathogens in Molecular Plant Pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A Unified Classification System for Eukaryotic Transposable Elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.W.; Proctor, R.H. Fusarium: Genomics, Molecular and Cellular Biology; Caister Academic Press: Poole, UK, 2013; p. 191. [Google Scholar] [CrossRef]
- Mahfooz, S.; Srivastava, A.; Srivastava, A.K.; Arora, D.K. A Comparative Analysis of Distribution and Conservation of Microsatellites in the Transcripts of Sequenced Fusarium Species and Development of Genic-SSR Markers for Polymorphism Analysis. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [Green Version]
- Wiemann, P.; Sieber, C.M.K.; von Bargen, K.W.; Studt, L.; Niehaus, E.M.; Espino, J.J.; Huß, K.; Michielse, C.B.; Albermann, S.; Wagner, D.; et al. Deciphering the Cryptic Genome: Genome-Wide Analyses of the Rice Pathogen Fusarium fujikuroi Reveal Complex Regulation of Secondary Metabolism and Novel Metabolites. PLOS Pathog. 2013, 9, e1003475. [Google Scholar] [CrossRef] [Green Version]
- Darnetty, T.; Darnetty, T.; Salleh, B. Toxigenicity of Fusarium Species in Gibberella fujikuroi Species Complex (GFSC) Associated with Stalk and Ear Rot Disease of Corn. Int. J. Phytopathol. 2013, 2, 147–154. [Google Scholar] [CrossRef]
- Ekwomadu, T.I.; Akinola, S.A.; Mwanza, M. Fusarium Mycotoxins, Their Metabolites (Free, Emerging, and Masked), Food Safety Concerns, and Health Impacts. Int. J. Environ. Res. Public Health 2021, 18, 1741. [Google Scholar] [CrossRef]
- Proctor, R.H.; Van Hove, F.; Susca, A.; Stea, G.; Busman, M.; van der Lee, T.; Waalwijk, C.; Moretti, A.; Ward, T.J. Birth, Death and Horizontal Transfer of the Fumonisin Biosynthetic Gene Cluster during the Evolutionary Diversification of Fusarium. Mol. Microbiol. 2013, 90, 290–306. [Google Scholar] [CrossRef] [PubMed]
- Waalwijk, C.; Lee, T.V.D.; Vries, I.D.; Hesselink, T.; Arts, J.; Kema, G.H.J. Synteny in toxigenic Fusarium Species: The fumonisin gene cluster and the mating type region as examples. Eur. J. Plant Pathol. 2004, 110, 533–544. [Google Scholar] [CrossRef]
- Gaffoor, I.; Trail, F. Characterization of Two Polyketide Synthase Genes Involved in Zearalenone Biosynthesis in Gibberellazeae. Appl. Environ. Microbiol. 2006, 72, 1793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blast2GO. High-Quality Functional Annotation Up and Running within No Time. 2016. Available online: http://Www.Blast2go.Com/B2ghome (accessed on 6 July 2020).
- Google Search. Available online: http://repeatmasker.org/RepeatMasker-open-4-0-9-p2.tar.gz (accessed on 6 July 2020).
- MISA—MIcroSAtellite Identification Tool. Available online: http://pgrc.ipk-gatersleben.de/misa (accessed on 6 July 2020).
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila Melanogaster Strain W1118; Iso-2; Iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [Green Version]
- SignalPv4.1 (SignalP,2020)-Google Search. Available online: https://www.google.com/search?q=SignalP+v4.1 (accessed on 6 July 2020).
- TargetP Version 1.1 (Targetp, 2020). Google Search. Available online: https://www.google.com/search?q=TargetP+version%201.1 (accessed on 1 August 2020).
- Phobius-Google Search 2020. Available online: https://www.google.com/search?q=Phobius (accessed on 6 July 2020).
- TMHMM. Server v. 2.0: Prediction of Transmembrane Helices in Proteins 2016. Available online: http://Www.Cbs.Dtu.Dk/Ser-vices/TMHMM (accessed on 1 August 2020).
- PredGPI. Prediction Server 2020. Available online: http://Gpcr.Biocomp.Unibo.It/Predgpi (accessed on 1 August 2020).
- WoLFPSORT. Protein Subcellular Localization Prediction 2020. Available online: https://Wolfpsort.Hgc.Jp (accessed on 1 August 2020).
- PHI-Base: A New Database for Pathogen Host Interactions. Available online: https://www.google.com/search?q=PHIbase%3A+a+new+database+for+pathogen+host+interactions (accessed on 1 August 2020).
- Carbohydrate-Active EnZYmes Database. Available online: http://Www.Cazy.Org (accessed on 1 August 2020).

| Measurement | Details |
|---|---|
| HQ filtered raw data | 8.6 Million reads |
| HQ filtered raw data (read count × read length) | 1.3 Gb |
| Draft genome size (Mb) | 42.91 Mb |
| Coverage | 98.50% |
| Number of scaffolds | 638 |
| Largest contig | 6.2 Mb |
| Average scaffold size | 0.07 Mb |
| N50 | 4.23 Mb |
| Gaps | 1198 |
| (G + C) content | 48.24% |
| Repeats | 0.88% |
| Protein-coding genes | 15,053 |
| Average gene length (bp) | 3.72 kb |
| Gene density | 37 gene/100 kb |
| Secretory proteins | 2058 |
| Repetitive Elements | Number of Elements | Length of Sequence (bp) | Percentage of Sequence (%) |
|---|---|---|---|
| Retroelements | 391 | 27,767 | 0.06 |
| Simple repeats | 6873 | 290,113 | 0.68 |
| DNA Transposons | 49 | 3674 | 0.01 |
| Small RNA | 276 | 25,751 | 0.06 |
| hAT-Charlie | 6 | 442 | 0.001 |
| TcMar-Tigger | 5 | 399 | 0.001 |
| Unclassified | 1 | 56 | - |
| LTR elements | 3 | 213 | - |
| ERVL-MaLRs | 1 | 97 | - |
| Total interspersed repeats | 0 | 20,823 | 0.05 |
| Low complexity | 826 | 41,634 | 0.10 |
| Features | Fusarium verticillioides Strains | |||||
|---|---|---|---|---|---|---|
| BIONCL4 | 7600 | BRIP14953 | BRIP53590 | NRRL20984 | Fv10027_t1 | |
| Geographic origin | India | USA | Australia | Australia | USA | Italy |
| Isolation source | Maize | Maize | Maize | Maize | Maize | Maize |
| Assembly level | Scaffold | Chromosome | Chromosome | Chromosome | Scaffold | Contig |
| Gene bank assembly accession | ------ | GCA_000149555.1 | GCA_003316975.2 | GCA_003316995.2 | GCA_013759275.1 | GCA_020882315.1 |
| Sequencing Method | Illumina HiSeq | shotgun sequencing | Illumina HiSeq | Illumina HiSeq | Illumina MiSeq | Illumina; Oxford Nanopore |
| Draft genome size (Mb) | 42.91 Mb | 41.84 | 42.54 | 42.29 | 41.92 | 44.65 |
| Coverage | 98.0x | - | 90.0x | 100.0x | 50.0x | 60.0x |
| Number of scaffolds | 638 | 37 | 255 | 258 | 857 | 21 |
| ScaffoldsN50(Mb) | 4.23 | 1.95 | 4.02 | 4.02 | 0.10 | 2.91 |
| GC content (%) | 48.24% | 48.68 | 48.15 | 48.26 | 48.80 | 47.90 |
| Protein-coding genes | 15,053 | 20,574 | 13,769 | 13,508 | - | - |
| Query ID | Identity (%) | Gene ID | Protein | Function (s) |
|---|---|---|---|---|
| 1754_g | 98.18 | 30058694 | Acetyltransferase | FUM1_Highly reducing polyketide synthase, fumonisin biosynthesis |
| 1746_g | 98.38 | 30058700 | Cytochrome P450 monooxygenase | FUM2_oxidoreductase activity, fumonisin biosynthesis |
| 1752_g | 99.71 | 30058695 | Cytochrome P450 | FUM6_Bifunctional_cytochrome_P450/NADPH--P450_reductase activity, fumonisin biosynthesis |
| 1751_g | 100 | 30058696 | Dehydrogenase | FUM7_oxidoreductase activity, fumonisin biosynthesis |
| 1750_g | 98.93 | 5357319 | Aminotransferase | FUM8_transferase activity, fumonisin biosynthesis |
| 7269_g | 34.92 | 30061262 | Acyl-CoA_synthetase | FUM10_fumonisin biosynthesis |
| 11438_g | 55.05 | 30065034 | Tricarboxylate transporter | FUM11_integral component of membrane, fumonisin biosynthesis |
| 1745_g | 99.46 | 30058701 | NAD dependent epimerase/dehydratase | FUM13_oxidoreductase activity, fumonisin biosynthesis |
| 1744_g | 97.45 | 30058702 | Non-ribosomal peptide synthetase | FUM14_ligase activity, fumonisin biosynthesis |
| 1742_g | 98.47 | 30058703 | Acyl-CoA_synthetase | FUM16_ligase activity, fumonisin biosynthesis |
| 10200_g | 50.40 | 30064824 | Sphingosine N-acyltransferase- like protein | FUM17_ integral component of membrane, fumonisin biosynthesis |
| 6947_g | 30.21 | 30061525 | Non reducing polyketide synthase ZEA1 | ZEA1_3-oxoacyl-[acyl-carrier-protein] synthase activity |
| 13719_g | 30.99 | 30066158 | Highly reducing polyketide synthase ZEA2 | ZEA2_3-oxoacyl-[acyl-carrier-protein] synthase activity, oxidoreductase activity zearalenone biosynthesis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navale, V.D.; Sawant, A.M.; Gowda, V.U.; Vamkudoth, K.R. Assembly, Annotation, and Comparative Whole Genome Sequence of Fusarium verticillioides Isolated from Stored Maize Grains. Pathogens 2022, 11, 810. https://doi.org/10.3390/pathogens11070810
Navale VD, Sawant AM, Gowda VU, Vamkudoth KR. Assembly, Annotation, and Comparative Whole Genome Sequence of Fusarium verticillioides Isolated from Stored Maize Grains. Pathogens. 2022; 11(7):810. https://doi.org/10.3390/pathogens11070810
Chicago/Turabian StyleNavale, Vishwambar D., Amol M. Sawant, Varun U. Gowda, and Koteswara Rao Vamkudoth. 2022. "Assembly, Annotation, and Comparative Whole Genome Sequence of Fusarium verticillioides Isolated from Stored Maize Grains" Pathogens 11, no. 7: 810. https://doi.org/10.3390/pathogens11070810






