IL-10-Secreting CD8+ T Cells Specific for Human Cytomegalovirus (HCMV): Generation, Maintenance and Phenotype
Abstract
1. Introduction
2. Materials and Methods
2.1. Donor Sample and Ethics Statement
2.2. Peripheral Blood Mononuclear Cell Isolation
2.3. HCMV ORF Peptide Mixes
2.4. ARIA Study Dual FluoroSpot Assays
2.5. Intracellular Cytokine Staining of CD8+ T cells
2.6. FluoroSpot Analysis of Longitudinal Kidney Transplant Samples
2.7. Phenotyping of IL-10-Secreting CD8+ T Cells
2.8. FluoroSpot Analysis of PD-1-Depleted CD8+ T Cells
2.9. FluoroSpot Analysis of Paired Bone Marrow and Peripheral Blood Mononuclear Cells
2.10. Statistics
3. Results
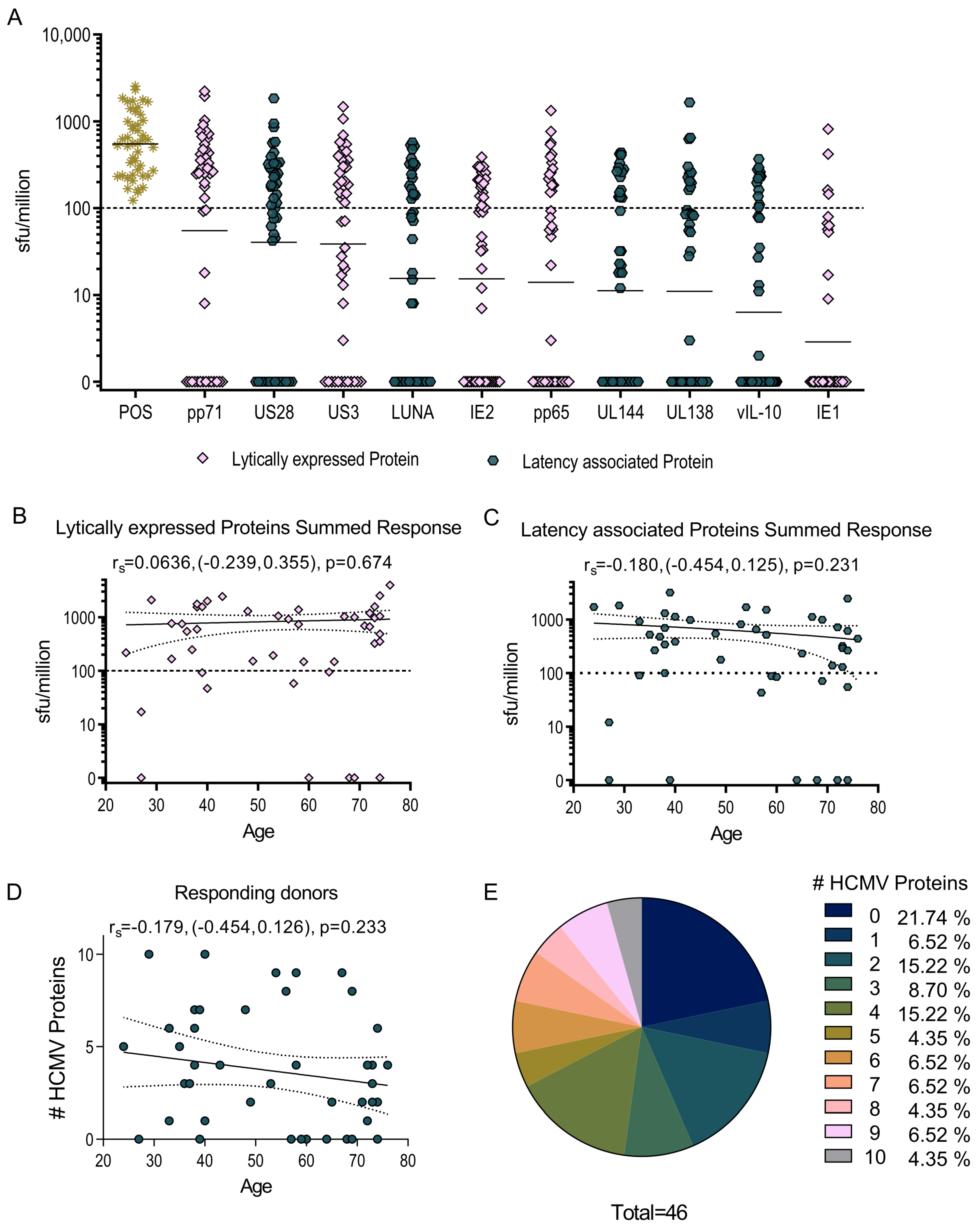
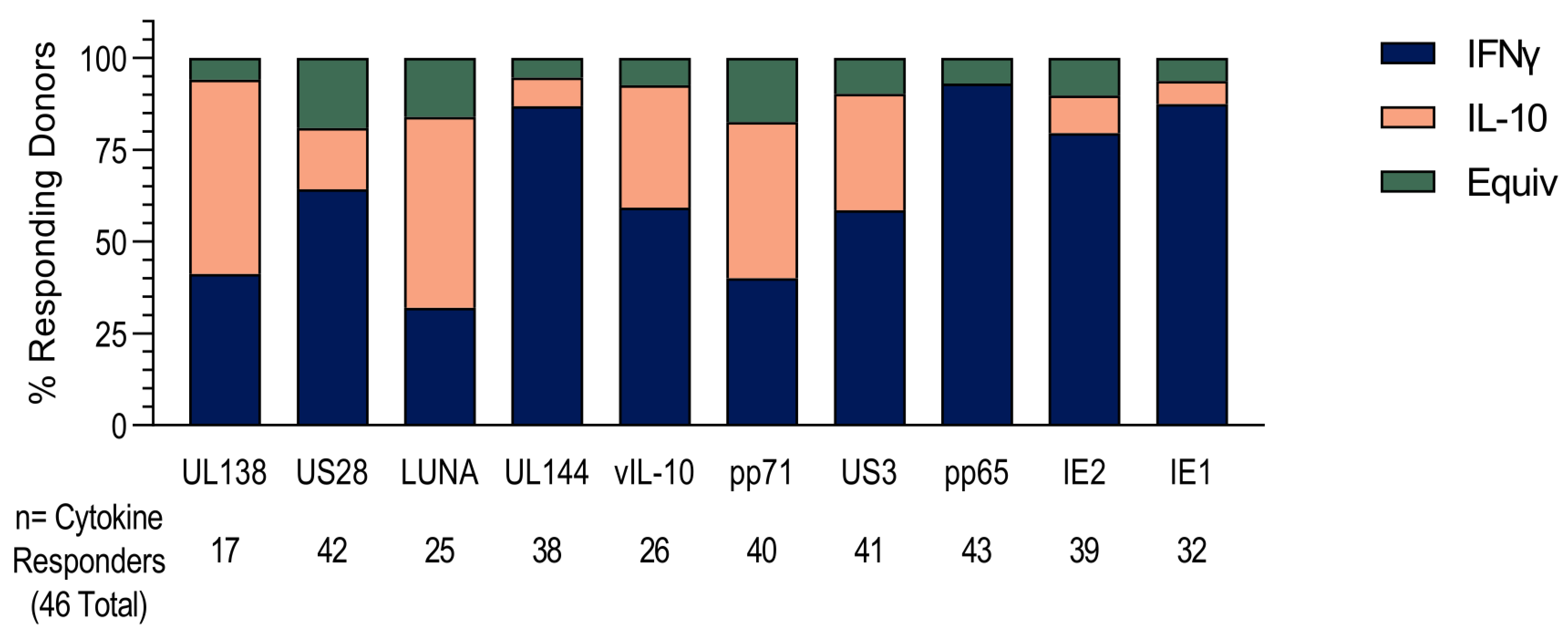
3.1. HCMV-Specific CD8+ T Cells Can Produce IL-10
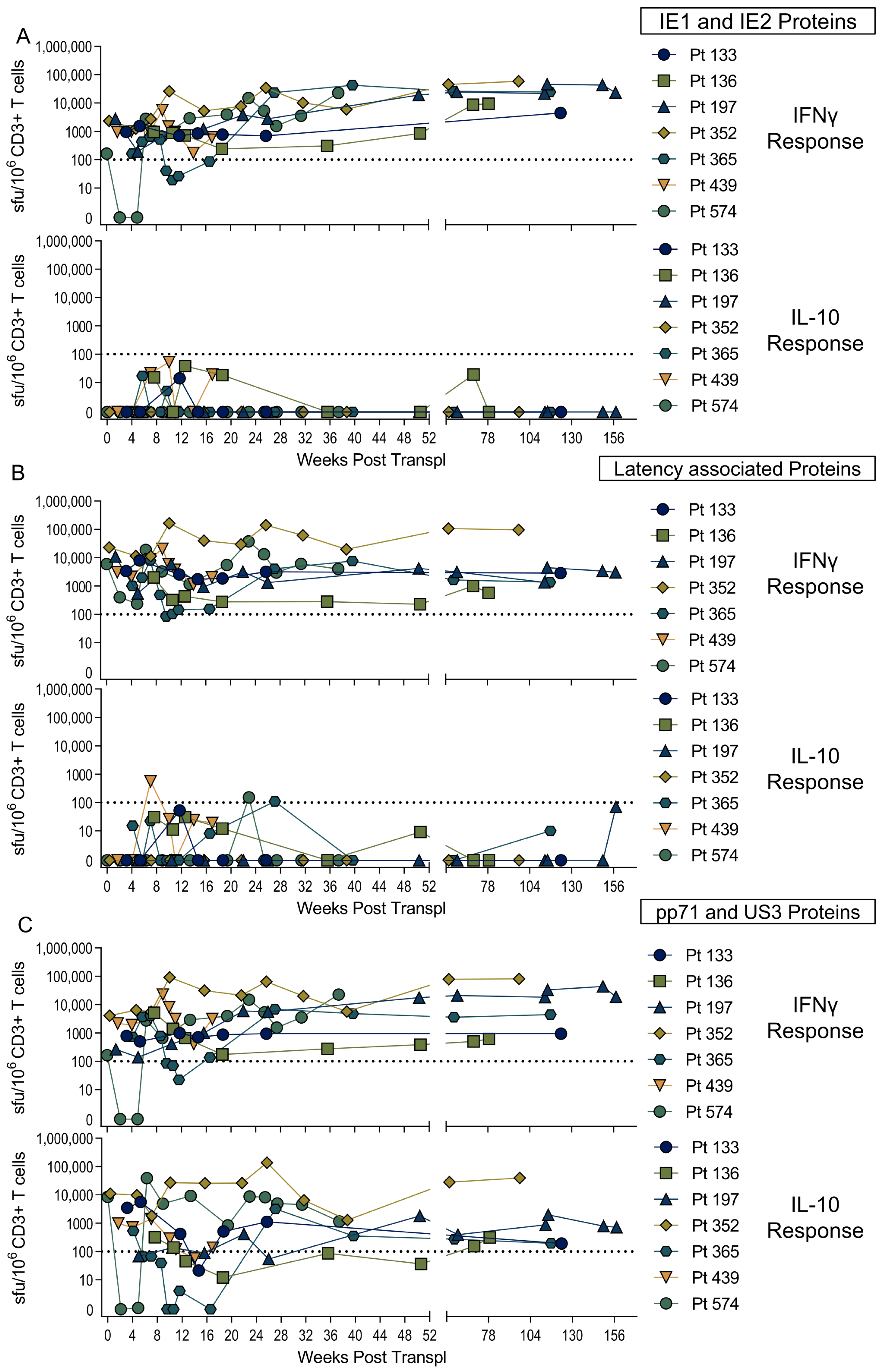
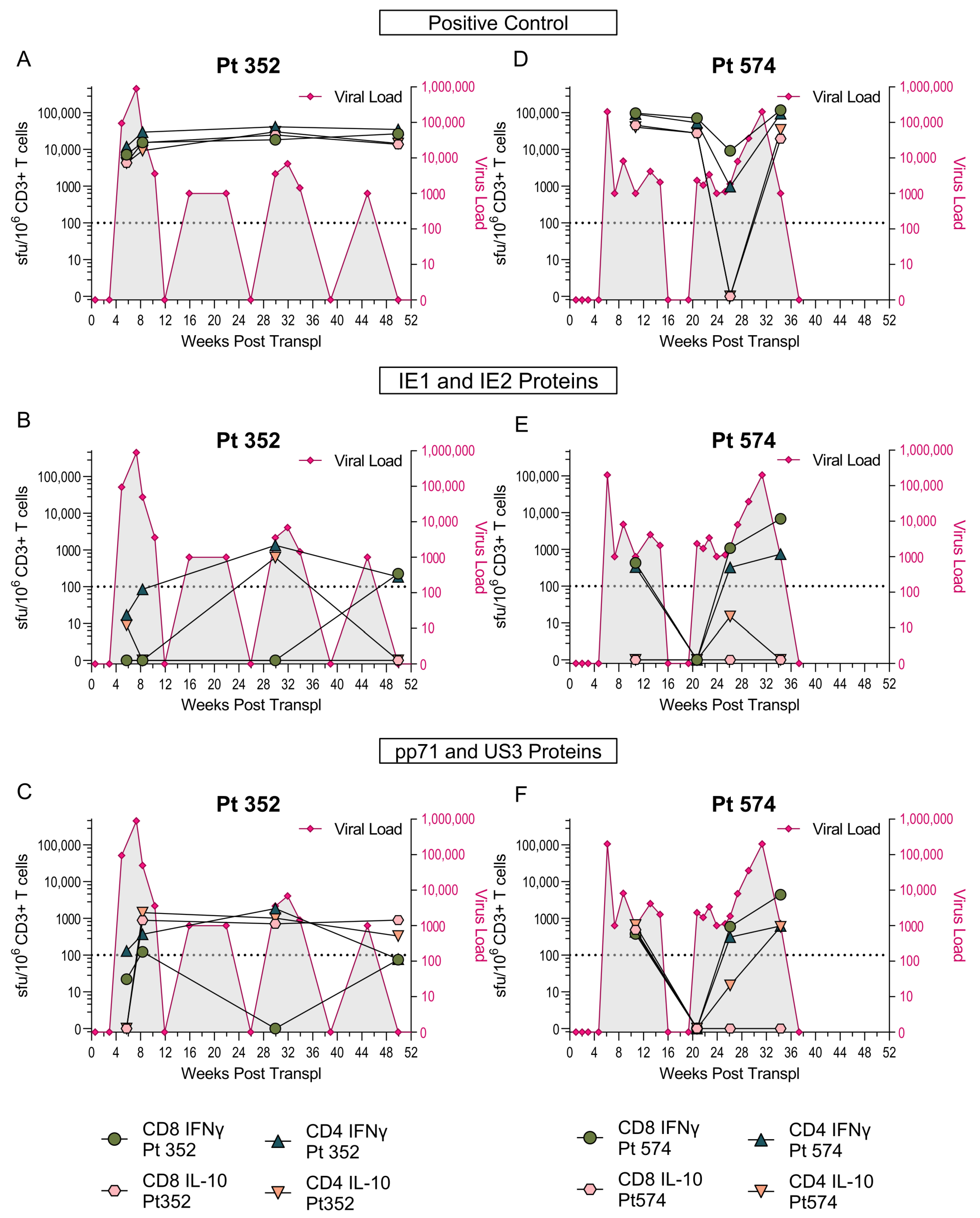
3.2. US3 and pp71 IL-10 CD8+ T cell Responses Arise Early in Primary Infection
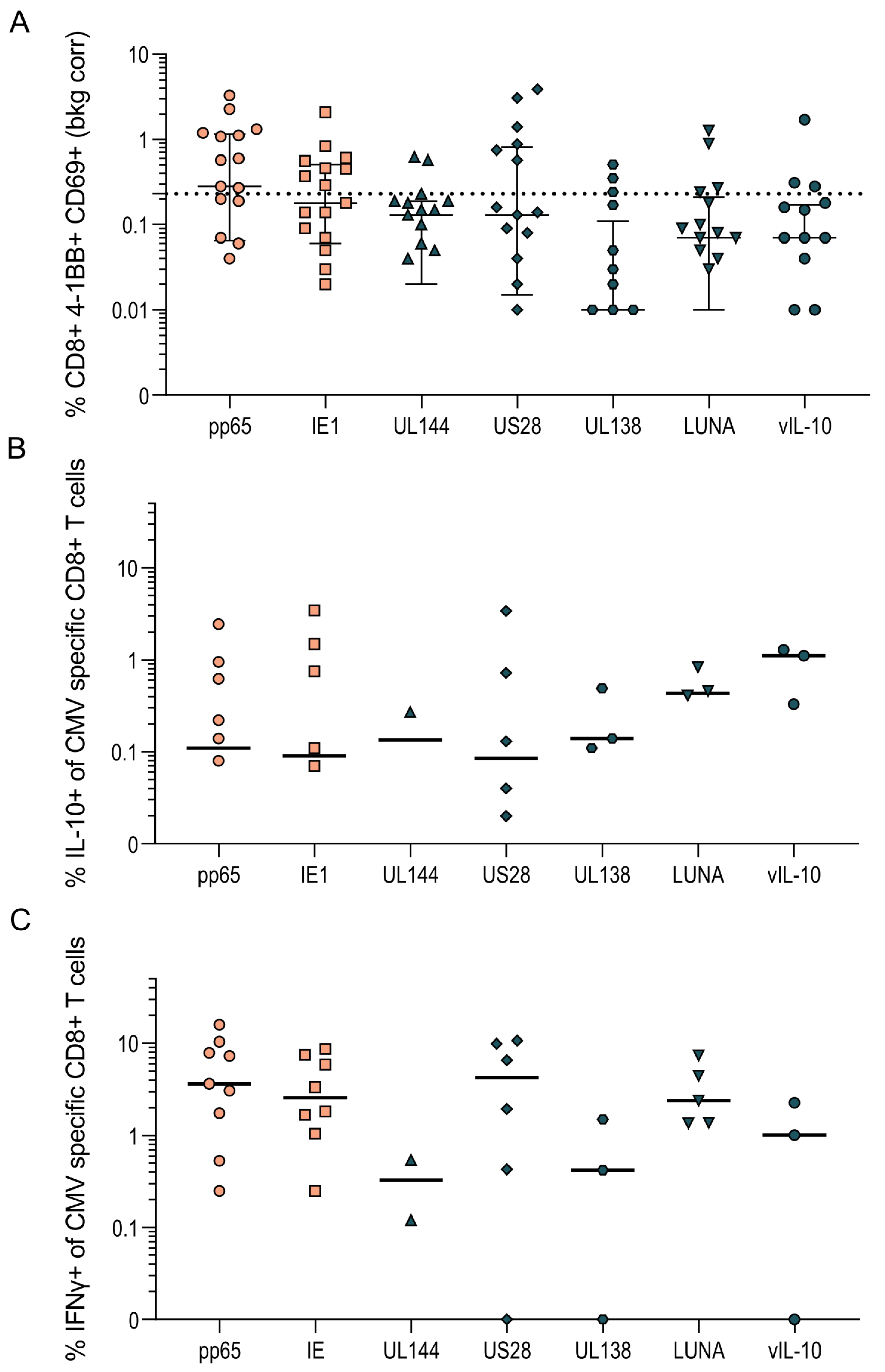
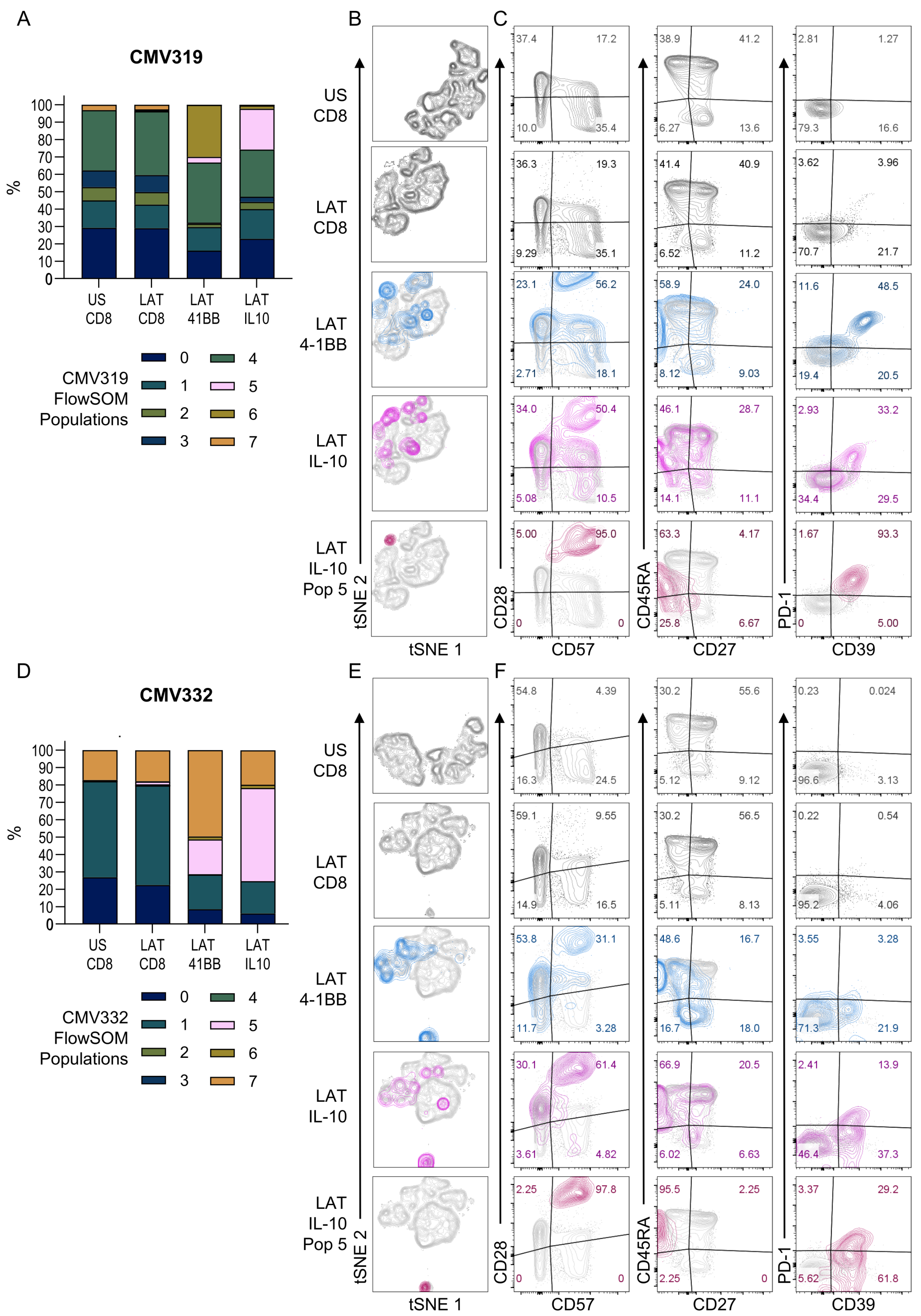
3.3. HCMV-Specific CD8+ IL-10-Secreting T Cells Are a Heterogeneous Population
3.4. Diverse HCMV-Specific IL-10 CD8+ T Cells May Be Tissue-Resident
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gandhi, M.K.; Khanna, R. Human cytomegalovirus: Clinical aspects, immune regulation, and emerging treatments. Lancet Infect. Dis. 2004, 4, 725–738. [Google Scholar] [CrossRef]
- Crough, T.; Khanna, R. Immunobiology of human cytomegalovirus: From bench to bedside. Clin. Microbiol. Rev. 2009, 22, 76–98. [Google Scholar] [CrossRef] [PubMed]
- Forte, E.; Zhang, Z.; Thorp, E.B.; Hummel, M. Cytomegalovirus Latency and Reactivation: An Intricate Interplay with the Host Immune Response. Front. Cell. Infect. Microbiol. 2020, 10, 130. [Google Scholar] [CrossRef] [PubMed]
- Shnayder, M.; Nachshon, A.; Krishna, B.; Poole, E.; Boshkov, A.; Binyamin, A.; Maza, I.; Sinclair, J.; Schwartz, M.; Stern-Ginossar, N. Defining the Transcriptional Landscape during Cytomegalovirus Latency with Single-Cell RNA Sequencing. Mbio 2018, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Caviness, K.; Buehler, J.; Smithey, M.; Nikolich-Zugich, J.; Goodrum, F. Transcriptome-wide characterization of human cytomegalovirus in natural infection and experimental latency. Proc. Natl. Acad. Sci. USA 2017, 114, E10586–E10595. [Google Scholar] [CrossRef]
- Sinclair, J.; Sissons, P. Latency and reactivation of human cytomegalovirus. J. Gen. Virol. 2006, 87, 1763–1779. [Google Scholar] [CrossRef]
- Picarda, G.; Benedict, C.A. Cytomegalovirus: Shape-Shifting the Immune System. J. Immunol. 2018, 200, 3881–3889. [Google Scholar] [CrossRef]
- Brodin, P.; Jojic, V.; Gao, T.; Bhattacharya, S.; Angel, C.J.; Furman, D.; Shen-Orr, S.; Dekker, C.L.; Swan, G.E.; Butte, A.J.; et al. Variation in the human immune system is largely driven by non-heritable influences. Cell 2015, 160, 37–47. [Google Scholar] [CrossRef]
- Jackson, S.E.; Mason, G.M.; Wills, M.R. Human cytomegalovirus immunity and immune evasion. Virus Res. 2011, 157, 151–160. [Google Scholar] [CrossRef]
- Khan, N.; Shariff, N.; Cobbold, M.; Bruton, R.; Ainsworth, J.A.; Sinclair, A.J.; Nayak, L.; Moss, P.A. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J. Immunol. 2002, 169, 1984–1992. [Google Scholar] [CrossRef]
- Sylwester, A.W.; Mitchell, B.L.; Edgar, J.B.; Taormina, C.; Pelte, C.; Ruchti, F.; Sleath, P.R.; Grabstein, K.H.; Hosken, N.A.; Kern, F.; et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 2005, 202, 673–685. [Google Scholar] [CrossRef]
- Van den Berg, S.P.H.; Pardieck, I.N.; Lanfermeijer, J.; Sauce, D.; Klenerman, P.; van Baarle, D.; Arens, R. The hallmarks of CMV-specific CD8 T-cell differentiation. Med. Microbiol. Immunol. 2019, 208, 365–373. [Google Scholar] [CrossRef]
- Jackson, S.E.; Mason, G.M.; Okecha, G.; Sissons, J.G.; Wills, M.R. Diverse specificities, phenotypes, and antiviral activities of cytomegalovirus-specific CD8+ T cells. J. Virol. 2014, 88, 10894–10908. [Google Scholar] [CrossRef]
- Jackson, S.E.; Sedikides, G.X.; Okecha, G.; Poole, E.L.; Sinclair, J.H.; Wills, M.R. Latent Cytomegalovirus (CMV) Infection Does Not Detrimentally Alter T Cell Responses in the Healthy Old, But Increased Latent CMV Carriage Is Related to Expanded CMV-Specific T Cells. Front. Immunol. 2017, 8, 733. [Google Scholar] [CrossRef]
- Hosie, L.; Pachnio, A.; Zuo, J.; Pearce, H.; Riddell, S.; Moss, P. Cytomegalovirus-Specific T Cells Restricted by HLA-Cw*0702 Increase Markedly with Age and Dominate the CD8+ T-Cell Repertoire in Older People. Front. Immunol. 2017, 8, 1776. [Google Scholar] [CrossRef]
- Gabanti, E.; Bruno, F.; Fornara, C.; Bernuzzi, S.; Lilleri, D.; Gerna, G. Polyfunctional analysis of human cytomegalovirus (HCMV)-specific CD4(+) and CD8 (+) memory T-cells in HCMV-seropositive healthy subjects following different stimuli. J. Clin. Immunol. 2014, 34, 999–1008. [Google Scholar] [CrossRef]
- Vieira Braga, F.A.; Hertoghs, K.M.; van Lier, R.A.; van Gisbergen, K.P. Molecular characterization of HCMV-specific immune responses: Parallels between CD8(+) T cells, CD4(+) T cells, and NK cells. Eur. J. Immunol. 2015, 45, 2433–2445. [Google Scholar] [CrossRef]
- Houldcroft, C.J.; Jackson, S.E.; Lim, E.Y.; Sedikides, G.X.; Davies, E.L.; Atkinson, C.; McIntosh, M.; Remmerswaal, E.B.M.; Okecha, G.; Bemelman, F.J.; et al. Assessing Anti-HCMV Cell Mediated Immune Responses in Transplant Recipients and Healthy Controls Using a Novel Functional Assay. Front. Cell. Infect. Microbiol. 2020, 10. [Google Scholar] [CrossRef]
- Choi, Y.J.; Kim, S.B.; Kim, J.H.; Park, S.H.; Park, M.S.; Kim, J.M.; Han, S.H.; Shin, E.C. Impaired polyfunctionality of CD8(+) T cells in severe sepsis patients with human cytomegalovirus reactivation. Exp. Mol. Med. 2017, 49, e382. [Google Scholar] [CrossRef][Green Version]
- Powers, C.; DeFilippis, V.; Malouli, D.; Fruh, K. Cytomegalovirus immune evasion. Curr. Top. Microbiol. Immunol. 2008, 325, 333–359. [Google Scholar]
- Wills, M.R.; Poole, E.; Lau, B.; Krishna, B.; Sinclair, J.H. The immunology of human cytomegalovirus latency: Could latent infection be cleared by novel immunotherapeutic strategies? Cell Mol. Immunol. 2015, 12, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Elder, E.G.; Krishna, B.A.; Williamson, J.; Lim, E.Y.; Poole, E.; Sedikides, G.X.; Wills, M.; O’Connor, C.M.; Lehner, P.J.; Sinclair, J. Interferon-Responsive Genes Are Targeted during the Establishment of Human Cytomegalovirus Latency. mBio 2019, 10, e02574-19. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.K.; Gottlieb, D.J.; Plachter, B.; Pepperl-Klindworth, S.; Avdic, S.; Cunningham, A.L.; Abendroth, A.; Slobedman, B. The role of the human cytomegalovirus UL111A gene in down-regulating CD4+ T-cell recognition of latently infected cells: Implications for virus elimination during latency. Blood 2009, 114, 4128–4137. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.E.; Chen, K.C.; Groves, I.J.; Sedikides, G.X.; Gandhi, A.; Houldcroft, C.J.; Poole, E.L.; Montanuy, I.; Mason, G.M.; Okecha, G.; et al. Latent Cytomegalovirus-Driven Recruitment of Activated CD4+ T Cells Promotes Virus Reactivation. Front. Immunol. 2021, 12, 657945. [Google Scholar] [CrossRef] [PubMed]
- Mason, G.M.; Poole, E.; Sissons, J.G.; Wills, M.R.; Sinclair, J.H. Human cytomegalovirus latency alters the cellular secretome, inducing cluster of differentiation (CD)4+ T-cell migration and suppression of effector function. Proc. Natl. Acad. Sci. USA 2012, 109, 14538–14543. [Google Scholar] [CrossRef]
- Tovar-Salazar, A.; Patterson-Bartlett, J.; Jesser, R.; Weinberg, A. Regulatory function of cytomegalovirus-specific CD4+CD27-CD28- T cells. Virology 2010, 398, 158–167. [Google Scholar] [CrossRef][Green Version]
- Schwele, S.; Fischer, A.M.; Brestrich, G.; Wlodarski, M.W.; Wagner, L.; Schmueck, M.; Roemhild, A.; Thomas, S.; Hammer, M.H.; Babel, N.; et al. Cytomegalovirus-specific regulatory and effector T cells share TCR clonality--possible relation to repetitive CMV infections. Am. J. Transpl. 2012, 12, 669–681. [Google Scholar] [CrossRef]
- Terrazzini, N.; Bajwa, M.; Vita, S.; Cheek, E.; Thomas, D.; Seddiki, N.; Smith, H.; Kern, F. A novel cytomegalovirus-induced regulatory-type T-cell subset increases in size during older life and links virus-specific immunity to vascular pathology. J. Infect. Dis. 2014, 209, 1382–1392. [Google Scholar] [CrossRef]
- Derhovanessian, E.; Chen, S.; Maier, A.B.; Hahnel, K.; de Craen, A.J.; Roelofs, H.; Westendorp, R.; Pawelec, G. CCR4+ Regulatory T Cells Accumulate in the Very Elderly and Correlate with Superior 8-Year Survival. J. Gerontol. A Biol. Sci. Med. Sci. 2015, 70, 917–923. [Google Scholar] [CrossRef]
- Clement, M.; Marsden, M.; Stacey, M.A.; Abdul-Karim, J.; Gimeno Brias, S.; Costa Bento, D.; Scurr, M.J.; Ghazal, P.; Weaver, C.T.; Carlesso, G.; et al. Cytomegalovirus-Specific IL-10-Producing CD4+ T Cells Are Governed by Type-I IFN-Induced IL-27 and Promote Virus Persistence. PLoS Pathog. 2016, 12, e1006050. [Google Scholar] [CrossRef]
- Mason, G.M.; Jackson, S.E.; Okecha, G.; Poole, E.; Sissons, J.G.; Sinclair, J.; Wills, M.R. Human cytomegalovirus latency-associated proteins elicit immune-suppressive IL-10 producing CD4(+) T cells. PLoS Pathog. 2013, 9, e1003635. [Google Scholar] [CrossRef]
- Namdari, H.; Hosseini, M.; Yazdanifar, M.; Farajifard, H.; Parvizpour, F.; Karamigolbaghi, M.; Hamidieh, A.A.; Rezaei, F. Protective and pathological roles of regulatory immune cells in human cytomegalovirus infection following hematopoietic stem cell transplantation. Rev. Med. Virol. 2021, 32, e2319. [Google Scholar] [CrossRef]
- Liston, A.; Aloulou, M. A fresh look at a neglected regulatory lineage: CD8+Foxp3+ Regulatory T cells. Immunol. Lett. 2022, 247, 22–26. [Google Scholar] [CrossRef]
- Niederlova, V.; Tsyklauri, O.; Chadimova, T.; Stepanek, O. CD8(+) Tregs revisited: A heterogeneous population with different phenotypes and properties. Eur. J. Immunol. 2021, 51, 512–530. [Google Scholar] [CrossRef]
- Lees, J.R. CD8+ T cells: The past and future of immune regulation. Cell. Immunol. 2020, 357, 104212. [Google Scholar] [CrossRef]
- Fenoglio, D.; Dentone, C.; Signori, A.; Di Biagio, A.; Parodi, A.; Kalli, F.; Nasi, G.; Curto, M.; Cenderello, G.; De Leo, P.; et al. CD8(+)CD28(-)CD127(lo)CD39(+) regulatory T-cell expansion: A new possible pathogenic mechanism for HIV infection? J. Allergy Clin. Immunol. 2018, 141, 2220–2233.e4. [Google Scholar] [CrossRef]
- Shi, Z.; Okuno, Y.; Rifa’i, M.; Endharti, A.T.; Akane, K.; Isobe, K.; Suzuki, H. Human CD8+CXCR3+ T cells have the same function as murine CD8+CD122+ Treg. Eur. J. Immunol. 2009, 39, 2106–2119. [Google Scholar] [CrossRef]
- Elrefaei, M.; Baker, C.A.; Jones, N.G.; Bangsberg, D.R.; Cao, H. Presence of suppressor HIV-specific CD8+ T cells is associated with increased PD-1 expression on effector CD8+ T cells. J. Immunol. 2008, 180, 7757–7763. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.Y.; Wherry, E.J.; Jin, B.; Xu, B.; Zou, Z.S.; Zhang, S.Y.; Li, B.S.; Wang, H.F.; Wu, H.; et al. Dynamic programmed death 1 expression by virus-specific CD8 T cells correlates with the outcome of acute hepatitis B. Gastroenterology 2008, 134, 1938–1949+1949.e1–3. [Google Scholar] [CrossRef]
- Wang, W.; Tong, Z.; Zhong, J.; Zhang, L.; Zhang, H.; Su, Y.; Gao, B.; Zhang, C. Identification of IL-10-secreting CD8(+)CD28(-)PD-1(+) regulatory T cells associated with chronic hepatitis C virus infection. Immunol. Lett. 2018, 202, 16–22. [Google Scholar] [CrossRef]
- Krishnan, A.; Zhou, W.; Lacey, S.F.; Limaye, A.P.; Diamond, D.J.; La Rosa, C. Programmed death-1 receptor and interleukin-10 in liver transplant recipients at high risk for late cytomegalovirus disease. Transpl. Infect. Dis. Off. J. Transplant. Soc. 2010, 12, 363–370. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, J.Y.; Zhang, Z.; Jin, B.; Zhang, S.Y.; Zhou, C.B.; Fu, J.L.; Wang, F.S. Cutting Edge: Programmed Death-1 Up-Regulation Is Involved in the Attrition of Cytomegalovirus-Specific CD8+ T Cells in Acute Self-Limited Hepatitis B Virus Infection. J. Immunol. 2008, 181, 3741–3744. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.E.; Sedikides, G.X.; Mason, G.M.; Okecha, G.; Wills, M.R. Human Cytomegalovirus (HCMV)-Specific CD4+ T Cells Are Polyfunctional and Can Respond to HCMV-Infected Dendritic Cells In Vitro. J. Virol. 2017, 91, 16. [Google Scholar] [CrossRef]
- Remmerswaal, E.B.; Havenith, S.H.; Idu, M.M.; van Leeuwen, E.M.; van Donselaar, K.A.; Ten Brinke, A.; van der Bom-Baylon, N.; Bemelman, F.J.; van Lier, R.A.; Ten Berge, I.J. Human virus-specific effector-type T cells accumulate in blood but not in lymph nodes. Blood 2012, 119, 1702–1712. [Google Scholar] [CrossRef] [PubMed]
- Boom, R.; Sol, C.; Weel, J.; Gerrits, Y.; Boer, M.D.; Dillen, P.W.-V. A Highly Sensitive Assay for Detection and Quantitation of Human Cytomegalovirus DNA in Serum and Plasma by PCR and Electrochemiluminescence. J. Clin. Microbiol. 1999, 37, 1489–1497. [Google Scholar] [CrossRef]
- Wills, M.R.; Carmichael, A.J.; Mynard, K.; Jin, X.; Weekes, M.P.; Plachter, B.; Sissons, J.G.P. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: Frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J. Virol. 1996, 70, 7569–7579. [Google Scholar] [CrossRef]
- Poole, E.; Walther, A.; Raven, K.; Benedict, C.A.; Mason, G.M.; Sinclair, J. The myeloid transcription factor GATA-2 regulates the viral UL144 gene during human cytomegalovirus latency in an isolate-specific manner. J. Virol. 2013, 87, 4261–4271. [Google Scholar] [CrossRef]
- Belkina, A.C.; Ciccolella, C.O.; Anno, R.; Halpert, R.; Spidlen, J.; Snyder-Cappione, J.E. Automated optimized parameters for T-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nat. Commun. 2019, 10, 5415. [Google Scholar] [CrossRef]
- Linderman, G.C.; Rachh, M.; Hoskins, J.G.; Steinerberger, S.; Kluger, Y. Fast interpolation-based t-SNE for improved visualization of single-cell RNA-seq data. Nat. Methods 2019, 16, 243–245. [Google Scholar] [CrossRef]
- Van Gassen, S.; Callebaut, B.; Van Helden, M.J.; Lambrecht, B.N.; Demeester, P.; Dhaene, T.; Saeys, Y. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytom. A 2015, 87, 636–645. [Google Scholar] [CrossRef]
- Jackson, S.E.; Sedikides, G.X.; Okecha, G.; Wills, M.R. Generation, maintenance and tissue distribution of T cell responses to human cytomegalovirus in lytic and latent infection. Med. Microbiol. Immunol. 2019, 208, 375–389. [Google Scholar] [CrossRef]
- Day, E.K.; Carmichael, A.J.; Ten Berge, I.J.M.; Waller, E.C.P.; Sissons, J.G.P.; Wills, M.R. Rapid CD8(+) T cell repertoire focusing and selection of high-affinity clones into memory following primary infection with a persistent human virus: Human Cytomegalovirus. J. Immunol. 2007, 179, 3203–3213. [Google Scholar] [CrossRef]
- Riou, R.; Bressollette-Bodin, C.; Boutoille, D.; Gagne, K.; Rodallec, A.; Lefebvre, M.; Raffi, F.; Senitzer, D.; Imbert-Marcille, B.M.; Retiere, C. Severe Symptomatic Primary Human Cytomegalovirus Infection despite Effective Innate and Adaptive Immune Responses. J. Virol. 2017, 91, 16. [Google Scholar] [CrossRef]
- Gerna, G.; Lilleri, D.; Fornara, C.; Bruno, F.; Gabanti, E.; Cane, I.; Furione, M.; Revello, M.G. Differential kinetics of human cytomegalovirus load and antibody responses in primary infection of the immunocompetent and immunocompromised host. J. Gen. Virol. 2015, 96, 360–369. [Google Scholar] [CrossRef][Green Version]
- Rentenaar, R.J.; Gamadia, L.E.; van DerHoek, N.; van Diepen, F.N.; Boom, R.; Weel, J.F.; Wertheim-van Dillen, P.M.; van Lier, R.A.; ten Berge, I.J. Development of virus-specific CD4(+) T cells during primary cytomegalovirus infection. J. Clin. Invest. 2000, 105, 541–548. [Google Scholar] [CrossRef]
- Gamadia, L.E.; Rentenaar, R.J.; Baars, P.A.; Remmerswaal, E.B.; Surachno, S.; Weel, J.F.; Toebes, M.; Schumacher, T.N.; ten Berge, I.J.; van Lier, R.A. Differentiation of cytomegalovirus-specific CD8(+) T cells in healthy and immunosuppressed virus carriers. Blood 2001, 98, 754–761. [Google Scholar] [CrossRef]
- Machicote, A.; Belen, S.; Baz, P.; Billordo, L.A.; Fainboim, L. Human CD8(+)HLA-DR(+) Regulatory T Cells, Similarly to Classical CD4(+)Foxp3(+) Cells, Suppress Immune Responses via PD-1/PD-L1 Axis. Front. Immunol. 2018, 9, 2788. [Google Scholar] [CrossRef]
- Kulpa, D.A.; Lawani, M.; Cooper, A.; Peretz, Y.; Ahlers, J.; Sekaly, R.P. PD-1 coinhibitory signals: The link between pathogenesis and protection. Semin. Immunol. 2013, 25, 219–227. [Google Scholar] [CrossRef]
- Parry, H.M.; Dowell, A.C.; Zuo, J.; Verma, K.; Kinsella, F.A.M.; Begum, J.; Croft, W.; Sharma-Oates, A.; Pratt, G.; Moss, P. PD-1 is imprinted on cytomegalovirus-specific CD4+ T cells and attenuates Th1 cytokine production whilst maintaining cytotoxicity. PLoS Pathog. 2021, 17, e1009349. [Google Scholar] [CrossRef]
- Di Rosa, F.; Pabst, R. The bone marrow: A nest for migratory memory T cells. Trends Immunol. 2005, 26, 360–366. [Google Scholar] [CrossRef]
- Clutton, G.; Yang, H.; Hancock, G.; Sande, N.; Holloway, C.; Angus, B.; von Delft, A.; Barnes, E.; Borrow, P.; Pellegrino, P.; et al. Emergence of a distinct HIV-specific IL-10-producing CD8+ T-cell subset with immunomodulatory functions during chronic HIV-1 infection. Eur. J. Immunol. 2013, 43, 2875–2885. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Q.; Wagner, W.M.; Zheng, W.; Wikby, A.; Remarque, E.J.; Pawelec, G. Dysfunctional CMV-specific CD8(+) T cells accumulate in the elderly. Exp. Gerontol. 2004, 39, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Olver, S.D.; Price, P. Contrasting phenotypes of liver-infiltrating leucocytes isolated from MCMV-infected BALB/c and C57BL/6 mice. Int. J. Exp. Pathol. 1998, 79, 33–46. [Google Scholar] [PubMed]
- Jost, N.H.; Abel, S.; Hutzler, M.; Sparwasser, T.; Zimmermann, A.; Roers, A.; Muller, W.; Klopfleisch, R.; Hengel, H.; Westendorf, A.M.; et al. Regulatory T cells and T-cell-derived IL-10 interfere with effective anti-cytomegalovirus immune response. Immunol. Cell. Biol. 2014, 92, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Gaddi, P.J.; Crane, M.J.; Kamanaka, M.; Flavell, R.A.; Yap, G.S.; Salazar-Mather, T.P. IL-10 Mediated Regulation of Liver Inflammation during Acute Murine Cytomegalovirus Infection. PLoS ONE 2012, 7, e42850. [Google Scholar] [CrossRef]
- Almanan, M.; Raynor, J.; Sholl, A.; Wang, M.; Chougnet, C.; Cardin, R.D.; Hildeman, D.A. Tissue-specific control of latent CMV reactivation by regulatory T cells. PLoS Pathog. 2017, 13, e1006507. [Google Scholar] [CrossRef]
- Sun, J.; Dodd, H.; Moser, E.K.; Sharma, R.; Braciale, T.J. CD4+ T cell help and innate-derived IL-27 induce Blimp-1-dependent IL-10 production by antiviral CTLs. Nat. Immunol. 2011, 12, 327–334. [Google Scholar] [CrossRef]
- Sun, J.; Madan, R.; Karp, C.L.; Braciale, T.J. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat. Med. 2009, 15, 277–284. [Google Scholar] [CrossRef]
- Palmer, E.M.; Holbrook, B.C.; Arimilli, S.; Parks, G.D.; Alexander-Miller, M.A. IFNgamma-producing, virus-specific CD8+ effector cells acquire the ability to produce IL-10 as a result of entry into the infected lung environment. Virology 2010, 404, 225–230. [Google Scholar] [CrossRef]
- Loebbermann, J.; Schnoeller, C.; Thornton, H.; Durant, L.; Sweeney, N.P.; Schuijs, M.; O’Garra, A.; Johansson, C.; Openshaw, P.J. IL-10 regulates viral lung immunopathology during acute respiratory syncytial virus infection in mice. PLoS ONE 2012, 7, e32371. [Google Scholar] [CrossRef]
- Elrefaei, M.; Burke, C.M.; Baker, C.A.; Jones, N.G.; Bousheri, S.; Bangsberg, D.R.; Cao, H. TGF-beta and IL-10 production by HIV-specific CD8+ T cells is regulated by CTLA-4 signaling on CD4+ T cells. PLoS ONE 2009, 4, e8194. [Google Scholar] [CrossRef]
- Accapezzato, D.; Francavilla, V.; Paroli, M.; Casciaro, M.; Chircu, L.V.; Cividini, A.; Abrignani, S.; Mondelli, M.U.; Barnaba, V. Hepatic expansion of a virus-specific regulatory CD8(+) T cell population in chronic hepatitis C virus infection. J. Clin. Invest. 2004, 113, 963–972. [Google Scholar] [CrossRef]
- Pangrazzi, L.; Reidla, J.; Carmona Arana, J.A.; Naismith, E.; Miggitsch, C.; Meryk, A.; Keller, M.; Krause, A.A.N.; Melzer, F.L.; Trieb, K.; et al. CD28 and CD57 define four populations with distinct phenotypic properties within human CD8(+) T cells. Eur. J. Immunol. 2019, 50, 363–379. [Google Scholar] [CrossRef]
- Smith, C.J.; Snyder, C.M. Inhibitory Molecules PD-1, CD73 and CD39 Are Expressed by CD8(+) T Cells in a Tissue-Dependent Manner and Can Inhibit T Cell Responses to Stimulation. Front. Immunol. 2021, 12, 704862. [Google Scholar] [CrossRef]
- Zangger, N.; Oxenius, A. T cell immunity to cytomegalovirus infection. Curr. Opin. Immunol. 2022, 77, 102185. [Google Scholar] [CrossRef]
- Reeves, M.B.; MacAry, P.A.; Lehner, P.J.; Sissons, J.G.; Sinclair, J.H. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. USA 2005, 102, 4140–4145. [Google Scholar] [CrossRef]
- Young, V.P.; Mariano, M.C.; Tu, C.C.; Allaire, K.M.; Avdic, S.; Slobedman, B.; Spencer, J.V. Modulation of the Host Environment by Human Cytomegalovirus with Viral Interleukin 10 in Peripheral Blood. J. Infect. Dis. 2017, 215, 874–882. [Google Scholar] [CrossRef]
- Pai, J.A.; Satpathy, A.T. High-throughput and single-cell T cell receptor sequencing technologies. Nat. Methods 2021, 18, 881–892. [Google Scholar] [CrossRef]
- Chang, C.X.; Tan, A.T.; Or, M.Y.; Toh, K.Y.; Lim, P.Y.; Chia, A.S.; Froesig, T.M.; Nadua, K.D.; Oh, H.L.; Leong, H.N.; et al. Conditional ligands for Asian HLA variants facilitate the definition of CD8+ T-cell responses in acute and chronic viral diseases. Eur. J. Immunol. 2013, 43, 1109–1120. [Google Scholar] [CrossRef]
- Haidar, G.; Boeckh, M.; Singh, N. Cytomegalovirus Infection in Solid Organ and Hematopoietic Cell Transplantation: State of the Evidence. J. Infect. Dis. 2020, 221 (Suppl. 1), S23–S31. [Google Scholar] [CrossRef]
- Lambe, G.; Mansukhani, D.; Khodaiji, S.; Shetty, A.; Rodrigues, C.; Kapadia, F. Immune Modulation and Cytomegalovirus Reactivation in Sepsis-induced Immunosuppression: A Pilot Study. Indian J. Crit. Care Med. 2022, 26, 53–61. [Google Scholar] [PubMed]
- Gatto, I.; Biagioni, E.; Coloretti, I.; Farinelli, C.; Avoni, C.; Caciagli, V.; Busani, S.; Sarti, M.; Pecorari, M.; Gennari, W.; et al. Cytomegalovirus blood reactivation in COVID-19 critically ill patients: Risk factors and impact on mortality. Intensive Care Med. 2022, 48, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Stranavova, L.; Pelak, O.; Svaton, M.; Hruba, P.; Fronkova, E.; Slavcev, A.; Osickova, K.; Maluskova, J.; Hubacek, P.; Fronek, J.; et al. Heterologous Cytomegalovirus and Allo-Reactivity by Shared T Cell Receptor Repertoire in Kidney Transplantation. Front. Immunol. 2019, 10, 2549. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jackson, S.E.; Sedikides, G.X.; Romashova, V.; Okecha, G.; Remmerswaal, E.B.M.; Bemelman, F.J.; Sinclair, J.H.; Wills, M.R. IL-10-Secreting CD8+ T Cells Specific for Human Cytomegalovirus (HCMV): Generation, Maintenance and Phenotype. Pathogens 2022, 11, 1530. https://doi.org/10.3390/pathogens11121530
Jackson SE, Sedikides GX, Romashova V, Okecha G, Remmerswaal EBM, Bemelman FJ, Sinclair JH, Wills MR. IL-10-Secreting CD8+ T Cells Specific for Human Cytomegalovirus (HCMV): Generation, Maintenance and Phenotype. Pathogens. 2022; 11(12):1530. https://doi.org/10.3390/pathogens11121530
Chicago/Turabian StyleJackson, Sarah E., George X. Sedikides, Veronika Romashova, Georgina Okecha, Ester B. M. Remmerswaal, Frederike J. Bemelman, John H. Sinclair, and Mark R. Wills. 2022. "IL-10-Secreting CD8+ T Cells Specific for Human Cytomegalovirus (HCMV): Generation, Maintenance and Phenotype" Pathogens 11, no. 12: 1530. https://doi.org/10.3390/pathogens11121530
APA StyleJackson, S. E., Sedikides, G. X., Romashova, V., Okecha, G., Remmerswaal, E. B. M., Bemelman, F. J., Sinclair, J. H., & Wills, M. R. (2022). IL-10-Secreting CD8+ T Cells Specific for Human Cytomegalovirus (HCMV): Generation, Maintenance and Phenotype. Pathogens, 11(12), 1530. https://doi.org/10.3390/pathogens11121530








