Transcriptome Analysis of Eggplant Root in Response to Root-Knot Nematode Infection
Abstract
:1. Introduction
2. Results
2.1. Analysis of RKN Infection in Eggplant Roots
2.2. Overview of RNA Sequencing
2.3. De Novo Assembly of Tor Transcriptome
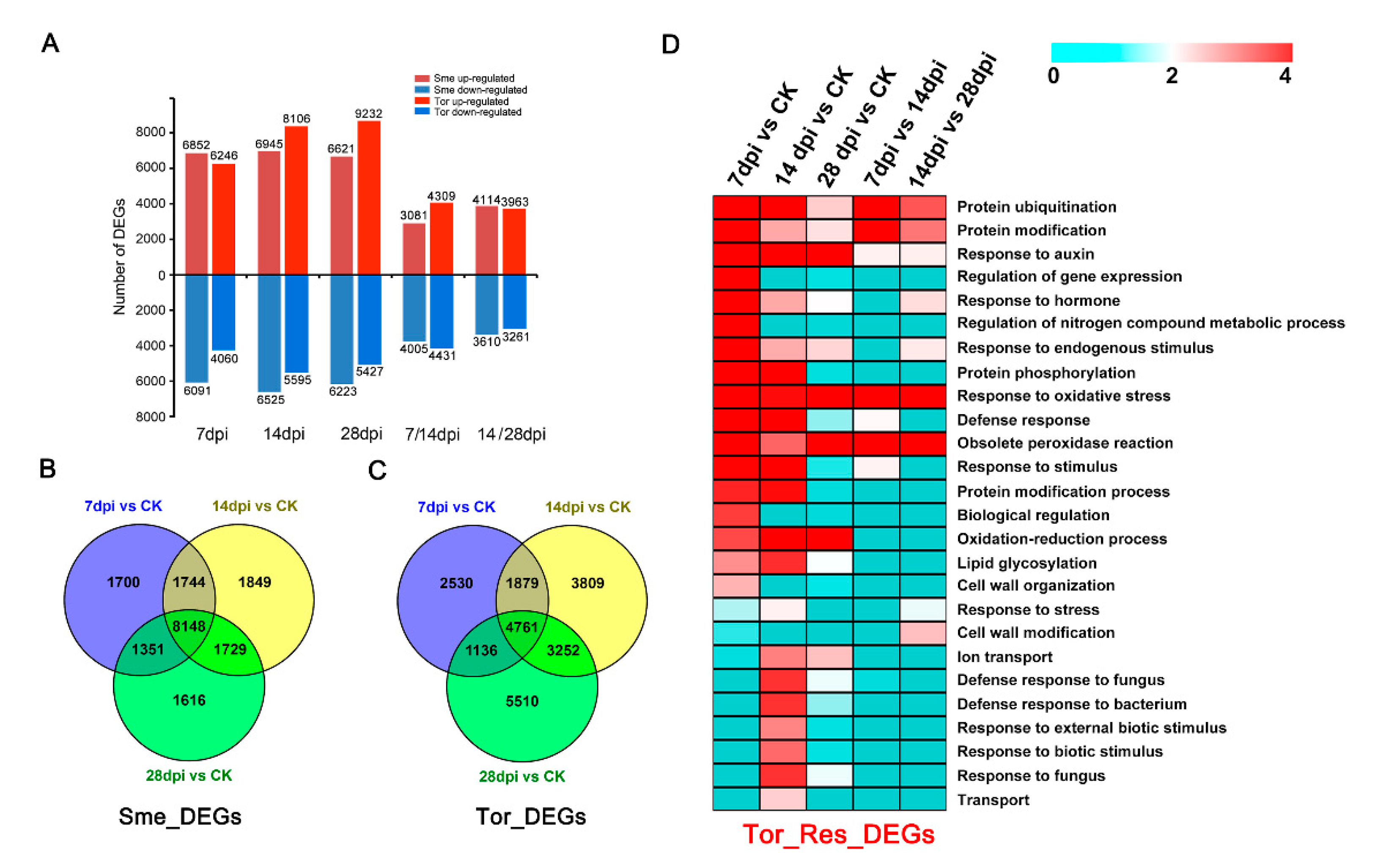
2.4. Identification of Differentially Expressed Genes under RKN Infection
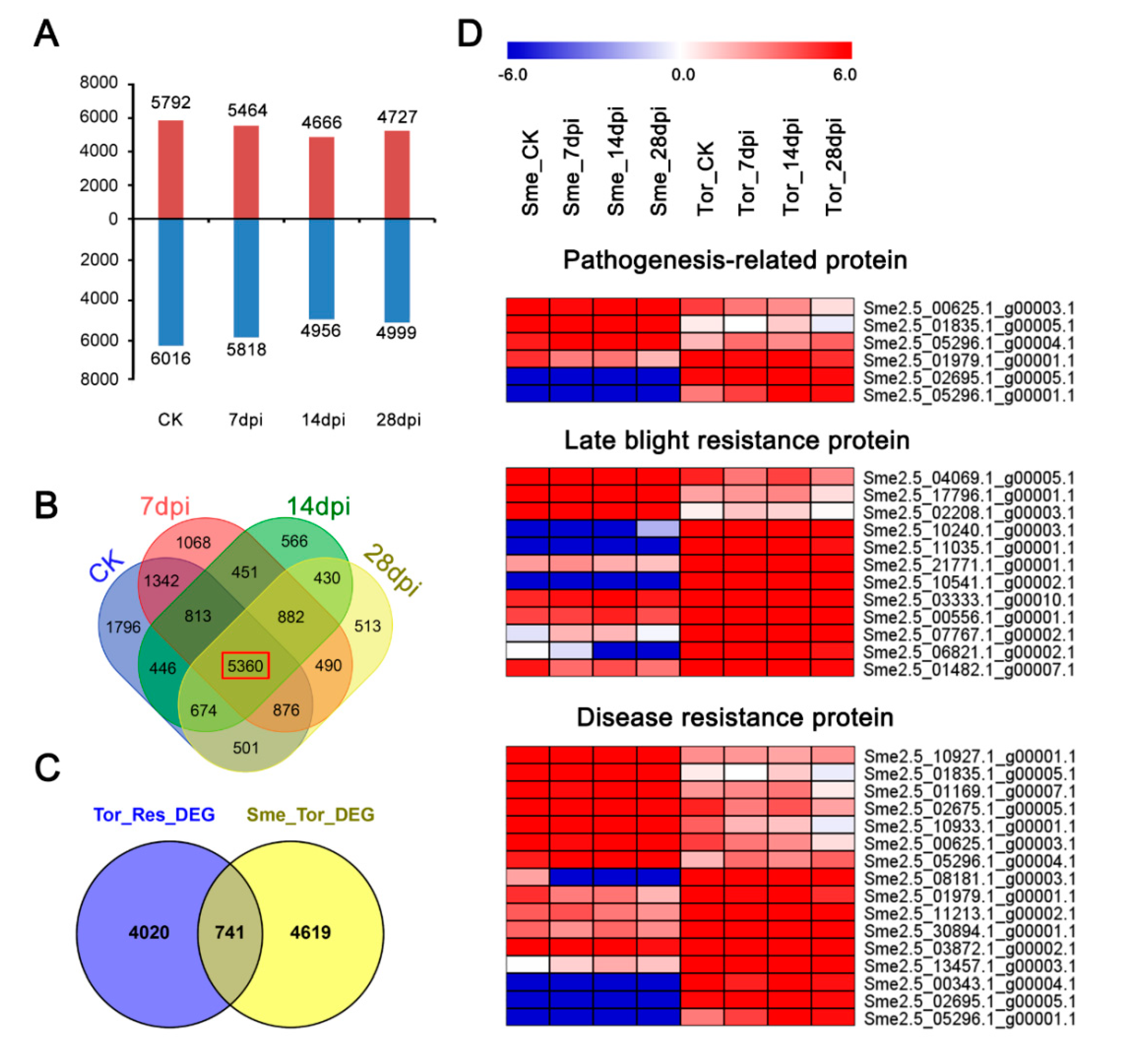
2.5. Comparisons of Transcriptomes between Sme and Tor Responses to RKN Infection
2.6. Specific Differentially Expressed Gene in Tor Responses to RKN Infection
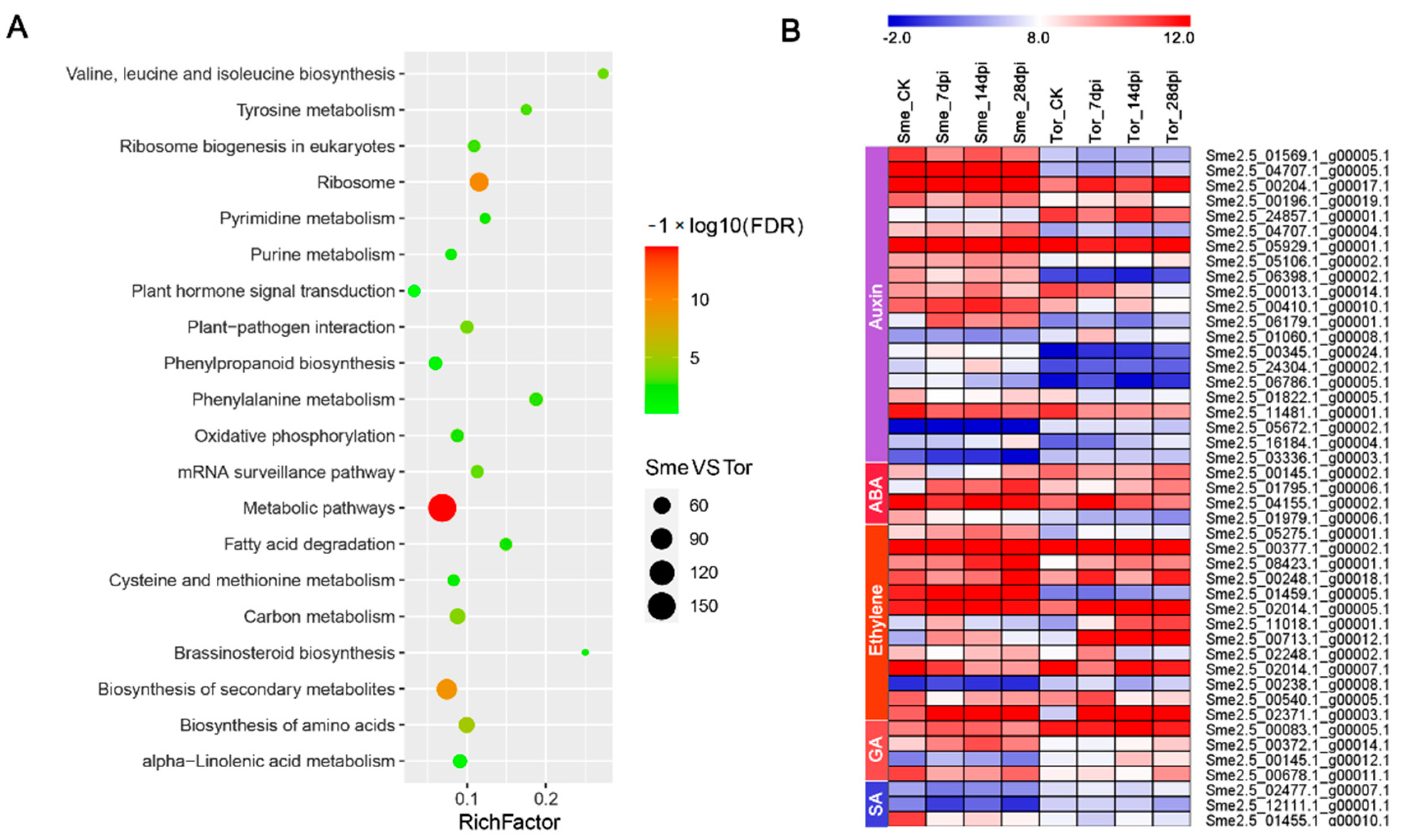
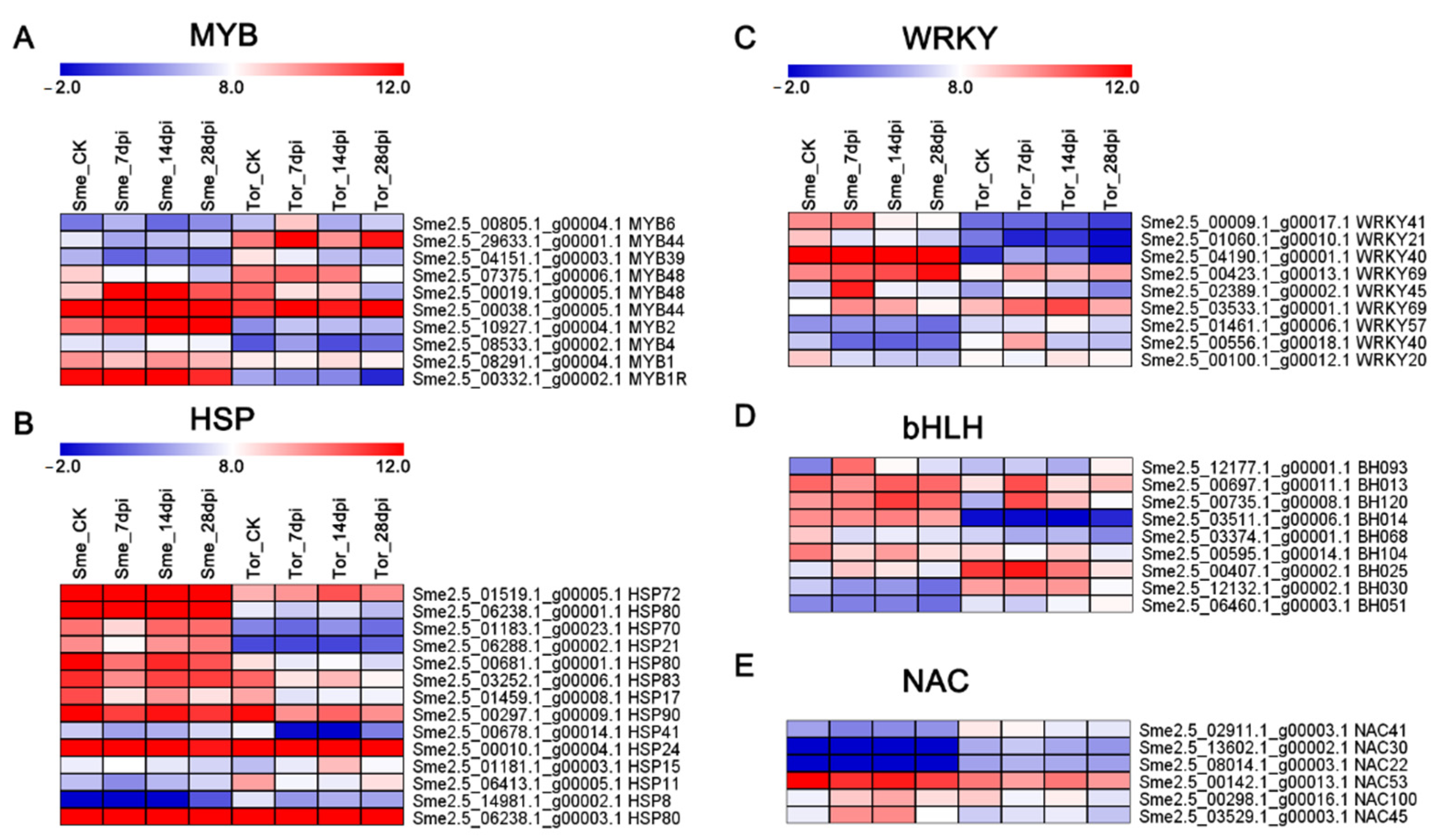
2.7. Analysis of Nematode-Responsive Hormone-Relative Genes and Transcription Factors
3. Discussion
4. Materials and Methods
4.1. Plant Material and Nematode Infection
4.2. RNA Extraction and Library Preparation for Transcriptome Sequencing
4.3. De Novo Assembly of S. torvum Transcriptome and Functional Annotation
4.4. Identification of Differentially Expressed Genes
4.5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
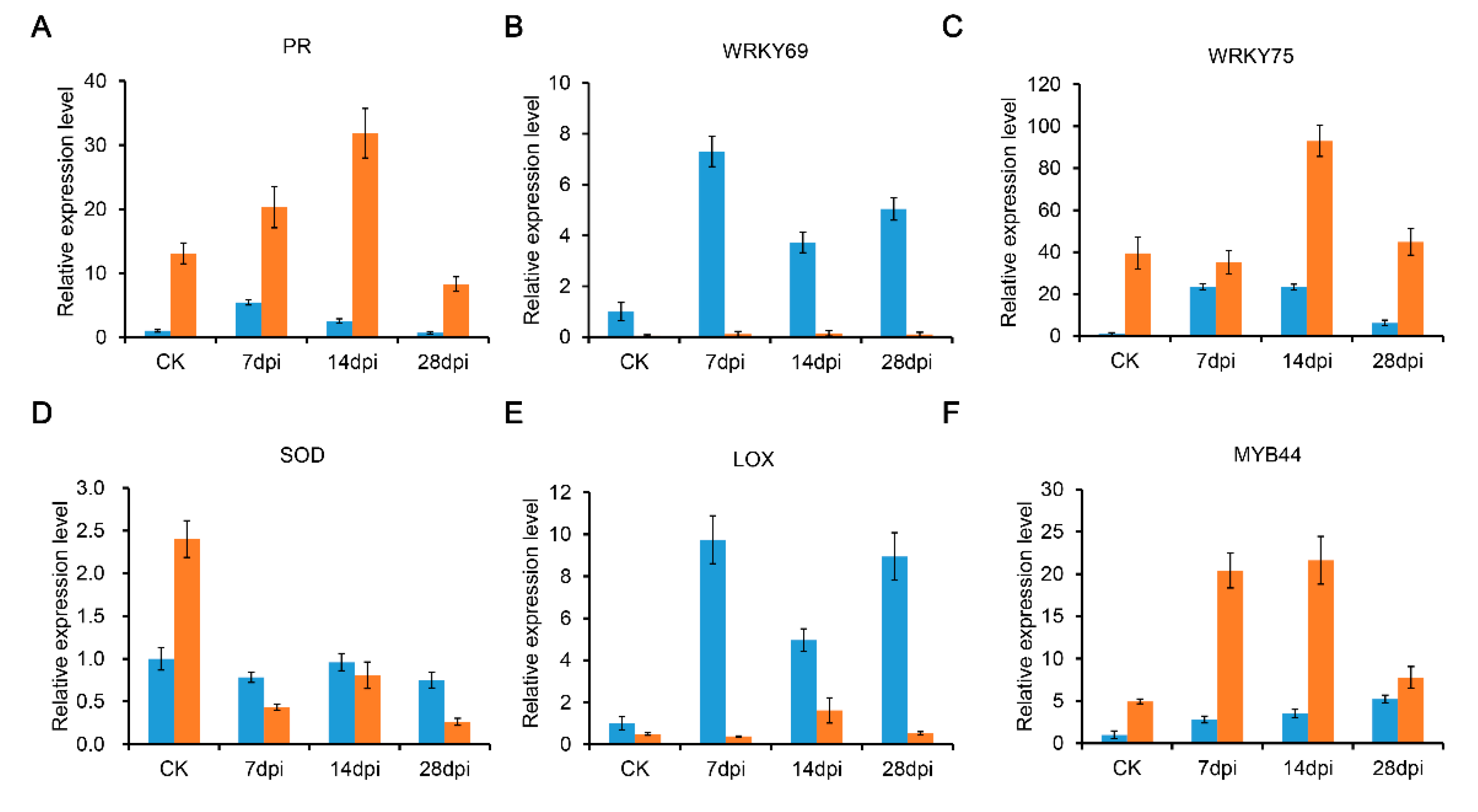
4.6. Quantitative Real-Time PCR (qRT-PCR) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, X.; Zhang, M.; Tan, J.; Huang, S.; Wang, C.; Zhang, H.; Tan, T. Comparative transcriptome analysis provides insights into molecular mechanisms for parthenocarpic fruit development in eggplant (Solanum melongena L.). PLoS ONE 2017, 12, e0179491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.J.; Wang, Y.M.; Long, L.K.; Lin, Y.; Pang, J.S.; Liu, B. Tomato rootstock effects on gene expression patterns in eggplant scions. Russian J. Plant Physiol. 2008, 55, 93–100. [Google Scholar] [CrossRef]
- Jiang, M.; Ren, L.; Lian, H.; Liu, Y.; Chen, H. Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 2016, 249, 46–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamosn, V.M.; Kumar, A. Nematode resistance in plants: The battle underground. Trends Genet. 2006, 22, 396–403. [Google Scholar] [CrossRef]
- Williamson, V.M. Root-knot nematode resistance genes in tomato and their potential for future use. Ann. Rev. Phytopathol. 1998, 36, 277–293. [Google Scholar] [CrossRef]
- Postnikova, O.A.; Hult, M.; Shao, J.; Skantar, A.; Nemchinov, L.G. Transcriptome analysis of resistant and susceptible Alfalfa cultivars infected with root-knot nematode Meloidogyne incognita. PLoS ONE 2015, 10, e0118269. [Google Scholar] [CrossRef]
- Ling, J.; Mao, Z.C.; Zhai, M.J.; Zeng, F.; Yang, Y.H.; Xie, B.Y. Transcriptome profiling of cucumis metuliferus infected by Meloidogyne incognita provides new insights into putative defense regulatory network in Cucurbitaceae. Sci. Rep. 2017, 7, 3544. [Google Scholar] [CrossRef]
- Barcala, M.; Garcia, A.; Cabrera, J.; Casson, S.; Lindsey, K.; Favery, B.; García-Casado, G.; Solano, R.; Fenoll, C.; Escobar, C. Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant J. 2010, 61, 698–712. [Google Scholar] [CrossRef]
- Yamaguchi, Y.L.; Suzuki, R.; Cabrera, J.; Nakagami, S.; Sagara, T.; Ejima, C.; Sano, R.; Aoki, Y.; Olmo, R.; Kurata, T.; et al. Root-knot and cyst nematodes activate procambium-associated genes in Arabidopsis roots. Front. Plant Sci. 2017, 8, 1195. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, D.; Petitot, A.S.; Grossi de Sá, M.; Nguyên, V.P.; de Almeida Engler, J.; Kyndt, T. Recent advances in understanding plant-nematode interactions in monocots. Adv. Bot. Res. 2015, 73, 189–219. [Google Scholar]
- Naseem, M.; Kaltdorf, M.; Dandekar, T. The nexus between growth and defence signaling: Auxin and cytokinin modulate plant immune response pathways. J. Exp. Bot. 2015, 66, 4885–4896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jammes, F.; Lecomete, P.; Almeida-Engler, J.; Bitton, F.; Martin-Magniette, M.L.; Renou, J.P.; Adad, P.; Favery, B. Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant J. 2005, 44, 447–458. [Google Scholar] [CrossRef]
- Oh, S.K.; Yi, S.Y.; Yu, S.H.; Moon, J.S. CaWRKY2, a chili pepper transcription factor, is rapidly induced by incompatible plant pathogens. Mol. Cell. 2006, 22, 58–64. [Google Scholar]
- Grunewald, W.; Karimi, M.; Wieczorek, K.; Cappelle, E.V.; Wischnitzki, E.; Grundler, F.; Inze, D.; Beeckman, T.; Gheysen, G. A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 2008, 148, 358–368. [Google Scholar] [CrossRef] [Green Version]
- Govindaraju, K.; Tamilselvan, S.; Kiruthiga, V.; Singaravelu, G. Biogenic silver nanoparticles by Solanum torvum and their promising antimicrobial activity. J. Biopestic. 2010, 3, 394–399. [Google Scholar]
- Hewezi, T.; Baum, T.J. Manipulation of plant cells by cyst and root-knot nematode effectors. MPMI 2013, 26, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.H.; Hu, J.H.; Huang, X.X.; Li, X.; Zhou, G.L.; Yan, Z.X. Comparative transcriptome analysis reveals heat-responsive genes in Chinese Cabbage (Brassica rapa ssp. chinensis). Front. Plant Sci. 2016, 7, 939. [Google Scholar]
- Bagnaresi, P.; Sala, T.; Irdani, T.; Scotto, C.; Lamontanara, A.; Beretta, M.; Rotino, G.L.; Sestili, S.; Cattivelli, L.; Sabatini, E. Solanum torvum responses to the root-knot nematode Meloidogyne incognita. BMC Genom. 2013, 14, 540. [Google Scholar] [CrossRef] [Green Version]
- Shukla, N.; Yadav, R.; Kaur, P.; Rasmussen, S.; Goel, S.; Agarwal, M.; Jagannath, A.; Gupta, R.; Kumar, A. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 2018, 19, 615–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, H.; Gheysen, G.; Denil, S.; Lindsey, K.; Topping, J.F.; Nahar, K.; Haegeman, A.; De Vos, W.H.; Trooskens, G.; Van Criekinge, W.; et al. Transcriptional analysis through RNA sequencing of giant cells induced by Meloidogyne graminicola in rice roots. J. Exp. Bot. 2013, 64, 3885–3898. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Cheng, Y.F.; Deng, C.; Ma, Y.; Wang, Z.W.; Chen, X.H.; Xue, L.B. Comparative transcriptome analysis of eggplant (Solanum melongena L.) and turkey berry (Solanum torvum Sw.): Phylogenomics and disease resistance analysis. BMC Genom. 2014, 15, 412. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.L.; Jang, G.; Um, T.; Kim, J.K.; Lee, J.S.; Yang, D.C. The soluble ABA receptor PYL8 regulates drought resistance by controlling ABA signaling in Arabidopsis. Plant Biotechnol. Rep. 2015, 9, 319–330. [Google Scholar]
- Zhou, X.H.; Liu, J.; Bao, S.Y.; Yang, Y.; Zhuang, Y. Molecular cloning and characterization of a wild eggplant Solanum aculeatissimum NBS-LRR gene, involved in plant resistance to Meloidogyne incognita. Int. J. Mol. Sci. 2018, 19, 583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyndt, T.; Denil, S.; Haegeman, A.; Trooskens, G.; Bauters, L.; Criekinge, W.V.; Meyer, T.D.; Gheysen, G. Transcriptional reprogramming by root knot and migratory nematode infection in rice. New Phytol. 2012, 196, 887–900. [Google Scholar] [CrossRef] [PubMed]
- De Young, B.J.; Inners, R.W. Plant NBS-LRR proteins in pathogen sensing and hose defense. Nat. Immunol. 2006, 7, 1243–1249. [Google Scholar]
- Hu, W.M.; Kingsbury, K.; Mishra, S.; DiGennaro, P. A comprehensive transcriptional profiling of pepper responses to root-knot nematode. Genes 2020, 11, 1507. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, D.; Shuang, L.; Xiao, D.; Xuan, Y.; Duan, Y.; Chen, L.; Wang, Y.; Liu, X.; Fan, H.; et al. Transcriptome analysis of rice roots in response to root-knot nematode infection. Int. J. Mol. Sci. 2020, 21, 848. [Google Scholar] [CrossRef] [Green Version]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-Encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, Y.; Zhou, X.H.; Wang, S.B. Genetic diversity of NBS-LRR class disease-resistance gene analogs in cultivated and wild eggplants. Plant Syst. Evol. 2012, 298, 1399–1406. [Google Scholar] [CrossRef]
- Djian-Caporalino, C.; Fazari, A.; Arguel, M.J.; Vernie, T.; VandeCasteele, C.; Faure, I.; Brunoud, G.; Pijarowski, L.; Palloix, A.; Lefebvre, V.; et al. Root-knot nematode (Meloidogyne spp.) Me resistance genes in pepper (Capsicum annuum L.) are clustered on the P9 chromosome. Theor. Appl. Genet. 2007, 114, 473–486. [Google Scholar] [CrossRef]
- Milligan, S.B.; Bodeau, J.; Yaghoobi, J.; Kaloshian, I.; Zabel, P.; Williamson, V.M. The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 1998, 10, 1307–1319. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.Y.; Kjemtrup-Lovelace, S.; Li, C.B.; Luo, Y.; Chen, L.P.; Song, B.H. Comparative RNA-seq analysis uncovers a complex regulatory network for soybean Cyst nematode resistance in wild soybean (Glycine soja). Sci. Rep. 2017, 7, 9699. [Google Scholar] [CrossRef] [Green Version]
- Grant, M.R.; Jones, J.D. Hormone (dis)harmony moulds plant health and disease. Science 2009, 324, 750–752. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Yin, C.C.; He, S.J.; Lu, X.; Zhang, W.K.; Lu, T.G.; Chen, S.Y. Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L.) seedlings. PLoS Genet. 2014, 10, e1004701. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defense response. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Flors, V.; Ton, J.; van Doorn, R.; Jakab, G.; Garcia-Agustin, P.; Mauch-Mani, B. Interplay between JA, SA, and ABA signaling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. 2007, 1, 81–92. [Google Scholar] [CrossRef]
- Shirano, Y.; Kachroo, P.; Shah, J.; Klessig, D.F. A Gain-of-Function mutation in an Arabidopsis Toll Interleukin 1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 2002, 14, 3149–3162. [Google Scholar] [CrossRef] [PubMed]
- Nahar, K.; Kyndt, T.; De Vleesschauwer, D.; Hofte, M.; Gheysen, G. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 2011, 157, 305–316. [Google Scholar] [CrossRef] [Green Version]
- Gousset, C.; Collonnier, C.; Mulya, K.; Mariska, I.; Rotino, G.L.; Besse, P.; Servaes, A.; Sihachakr, D. Solanum torvum, as a useful source of resistance against bacterial and fungal diseases for improvement of eggplant (S. melongena L.). Plant Sci. 2005, 168, 319–327. [Google Scholar] [CrossRef]
- Aribaud, M.; Jégo, S.; Wicker, E.; Fock, I. Ralstonia solanacearum induces soluble amine-oxidase activity in Solanum torvum stem calli. Plant Physiol. Biochem. 2010, 48, 787–796. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agriculture community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.Z.; Huang, J.J.; Ding, Y.; Wu, J.M.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L.P. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


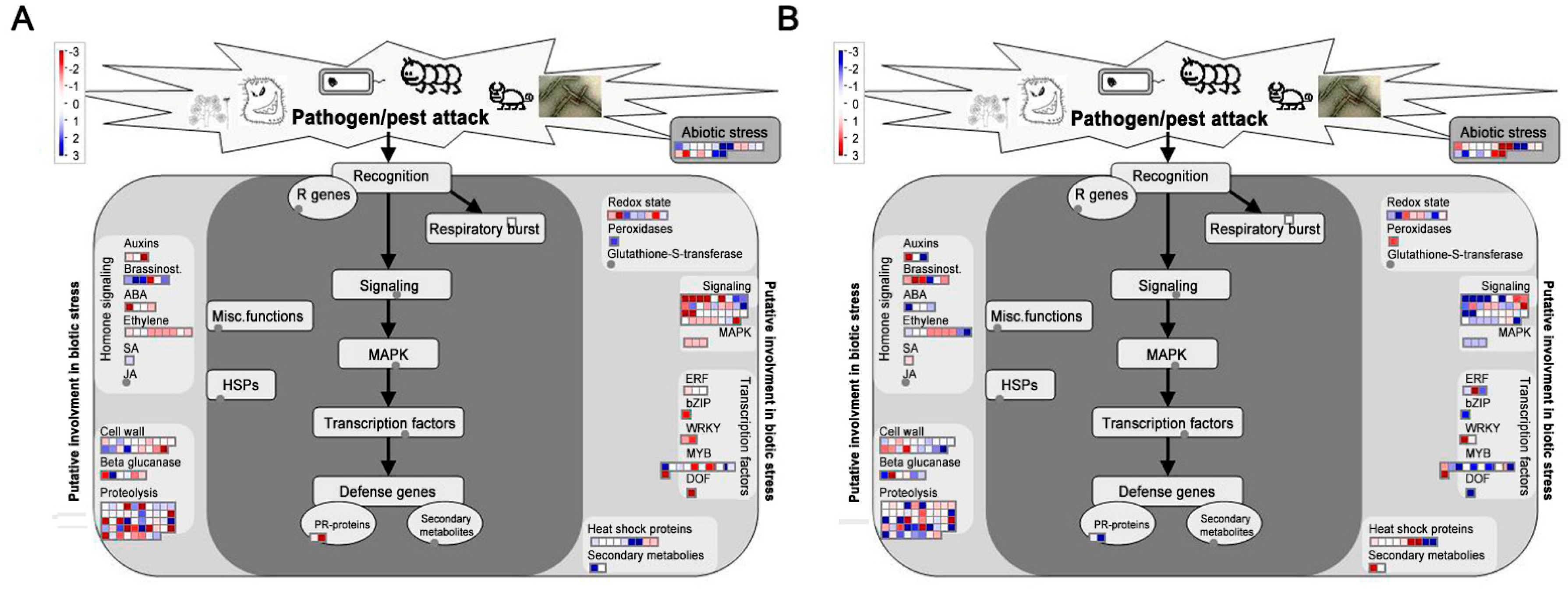
| Gene ID | Alias | CK a | 7 dpi | 14 dpi | 28 dpi | Annotation |
|---|---|---|---|---|---|---|
| Sme2.5_00077.1_g00005.1 | RPP13 | 1.73 | 1.13 | 0.68 | 1.58 | Disease resistance protein RPP13 |
| Sme2.5_03872.1_g00001.1 | RGA1 | 0.61 | 0.32 | 0.63 | 0.37 | Disease resistance protein RGA1 |
| Sme2.5_03872.1_g00002.1 | RGA4 | 0.39 | 0.25 | 0.21 | 0.26 | Disease resistance protein RGA4 |
| Sme2.5_00130.1_g00024.1 | RPM1 | 0.38 | 1.47 | 0.13 | 0.06 | Disease resistance protein RPM1 |
| Sme2.5_00345.1_g00018.1 | RPP8 | 0.37 | 0.84 | 0.64 | 0.33 | Disease resistance protein RPP8 |
| Sme2.5_07918.1_g00005.1 | RPM1 | 7.79 | 2.74 | 3.66 | 3.23 | Disease resistance protein RPM1 |
| Sme2.5_02954.1_g00001.1 | RPV1 | 0.98 | 2.20 | 3.86 | 2.65 | Disease resistance protein RPV1 |
| Sme2.5_05922.1_g00005.1 | RPM1 | 0.07 | 0.09 | 0.03 | 0.10 | Disease resistance RPM1 |
| Sme2.5_10927.1_g00001.1 | RGA4 | 7.92 | 4.52 | 5.64 | 6.58 | Disease resistance protein RGA4 |
| Sme2.5_06008.1_g00004.1 | RGA3 | 0.56 | 0.10 | 0.08 | 0.10 | Disease resistance protein RGA3 |
| Sme2.5_16574.1_g00002.1 | R13L4 | 0.58 | 0.87 | 1.20 | 1.34 | Disease resistance RPP13-like protein 4 |
| Sme2.5_13369.1_g00002.1 | RGA4 | 0.20 | 0.28 | 0.11 | 0.26 | Disease resistance protein RGA4 |
| Sme2.5_19803.1_g00001.1 | R13L4 | 0.88 | 0.29 | 0.27 | 1.14 | Disease resistance RPP13-like protein 4 |
| Gene ID | Alias | CK a | 7 dpi | 14 dpi | 28 dpi | Annotation |
|---|---|---|---|---|---|---|
| Sme2.5_00057.1_g00028.1 | PR1 | 2.651 | 1.797 | 1.175 | 1.868 | Pathogenesis-related protein 1 |
| Sme2.5_05922.1_g00005.1 | RPM1 | 0.071 | 0.093 | 0.028 | 0.096 | Disease resistance protein RPM1 |
| Sme2.5_03682.1_g00009.1 | DRL27 | 0.258 | 0.047 | 0.144 | 0.292 | Disease resistance protein DRL27 |
| Sme2.5_09833.1_g00003.1 | GSO2 | 0.865 | 4.465 | 0.425 | 0.399 | LRR receptor-like serine/threonine- |
| protein kinase GSO2 | ||||||
| Sme2.5_00365.1_g00011.1 | CML16 | 0.135 | 0.140 | 0.169 | 0.100 | Calcium-binding protein CML16 |
| Sme2.5_02438.1_g00002.1 | CDPK8 | 0.238 | 0.222 | 0.224 | 0.435 | Calcium-dependent protein kinase 8 |
| Sme2.5_00959.1_g00002.1 | CDPK2 | 3.369 | 0.940 | 0.942 | 1.091 | Calcium-dependent protein kinase 2 |
| Sme2.5_00572.1_g00003.1 | CNG18 | 0.304 | 0.340 | 0.241 | 0.289 | Cyclic nucleotide-gated ion channel 18 |
| Sme2.5_00629.1_g00003.1 | MRK7 | 5.936 | 12.89 | 8.233 | 9.426 | Mitogen-activated protein kinase kinase 7 |
| Sme2.5_00861.1_g00010.1 | MRK3 | 3.917 | 4.601 | 5.035 | 7.191 | Mitogen-activated protein kinase kinase 3 |
| Sme2.5_01071.1_g00003.1 | WRKY22 | 1.240 | 2.136 | 2.084 | 2.632 | WRKY transcription factor 22 |
| Sme2.5_00297.1_g00009.1 | HSP90 | 1.240 | 2.136 | 2.084 | 2.632 | Heat shock 90 kDa protein |
| Gene ID | CK a | 7 dpi | 14 dpi | 28 dpi | Annotation |
|---|---|---|---|---|---|
| Sme2.5_00236.1_g00007.1 | 6.104 | 1.235 | 1.871 | 2.275 | Chitin-inducible gibberellin-responsive protein |
| Sme2.5_18829.1_g00001.1 | 111.4 | 72.32 | 4.060 | 2.670 | Basic endochitinase A |
| Sme2.5_03222.1_g00007.1 | 11.86 | 65.26 | 26.70 | 74.93 | Endochitinase 1 |
| Sme2.5_00287.1_g00007.1 | 1.346 | 16.19 | 2.376 | 3.592 | Wound-induced protein |
| Sme2.5_03391.1_g00004.1 | 1.911 | 5.025 | 2.275 | 0.807 | Lignin-forming anionic peroxidase |
| Sme2.5_03391.1_g00006.1 | 3.010 | 2.131 | 3.572 | 1.007 | Lignin-forming anionic peroxidase |
| Sme2.5_00295.1_g00005.1 | 0.679 | 0.189 | 0.026 | 0.394 | Laccase-3 |
| Sme2.5_00135.1_g00019.1 | 7.591 | 2.562 | 2.642 | 2.476 | Xyloglucan endotransglucosylase/hydrolase 16 |
| Sme2.5_03583.1_g00006.1 | 1.745 | 1.184 | 0.443 | 0.226 | Xyloglucan endotransglucosylase/hydrolase 2 |
| Sme2.5_00498.1_g00013.1 | 0.031 | 0.052 | 0.410 | 0.000 | Xyloglucan endotransglucosylase/hydrolase 26 |
| Sme2.5_00096.1_g00018.1 | 8.966 | 1.924 | 1.952 | 2.720 | Xyloglucan endotransglucosylase/hydrolase 25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Zhang, H.; Tan, J.; Huang, S.; Chen, X.; Jiang, D.; Xiao, X. Transcriptome Analysis of Eggplant Root in Response to Root-Knot Nematode Infection. Pathogens 2021, 10, 470. https://doi.org/10.3390/pathogens10040470
Zhang M, Zhang H, Tan J, Huang S, Chen X, Jiang D, Xiao X. Transcriptome Analysis of Eggplant Root in Response to Root-Knot Nematode Infection. Pathogens. 2021; 10(4):470. https://doi.org/10.3390/pathogens10040470
Chicago/Turabian StyleZhang, Min, Hongyuan Zhang, Jie Tan, Shuping Huang, Xia Chen, Daohong Jiang, and Xueqiong Xiao. 2021. "Transcriptome Analysis of Eggplant Root in Response to Root-Knot Nematode Infection" Pathogens 10, no. 4: 470. https://doi.org/10.3390/pathogens10040470






