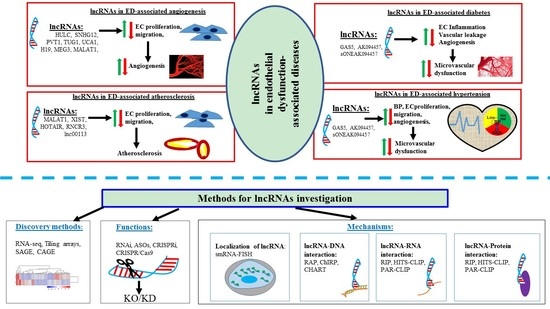
A Brief Overview of lncRNAs in Endothelial Dysfunction-Associated Diseases: From Discovery to Characterization
Abstract
:1. Introduction
2. lncRNAs in the Regulation of Gene Expression
3. Functional Roles of lncRNAs in Endothelial Dysfunction
4. Methods for Studying lncRNA: From Discovery to Function
4.1. Methods to Identify/Discover lncRNAs
4.1.1. Tiling Arrays
4.1.2. Serial Analysis of Gene Expression (SAGE)
4.1.3. Cap Analysis of Gene Expression (CAGE)
4.1.4. RNA Sequencing (RNA-Seq)
4.2. Methods to Study the Functional Roles of lncRNAs
4.2.1. RNA Interference (RNAi)
4.2.2. Antisense Oligonucleotides (ASOs)
4.2.3. CRISPR/Cas System
4.2.4. CRISPR Interference (CRISPRi)
4.3. Different Methods to Unveil the Functional Mechanisms of lncRNAs
4.3.1. Localization of lncRNA: Single-Molecule RNA Fluorescence In Situ Hybridization (smRNA-FISH)
4.3.2. Techniques for Investigating lncRNA-DNA Interaction
Capture Hybridization Analysis of RNA Target (CHART)
Chromatin Isolation by RNA Purification (ChIRP)
RNA Antisense Purification (RAP)
4.3.3. Techniques for Investigating lncRNA-RNA Interaction
RNA Antisense purification Followed by RNA Sequencing (RAP-RNA)
Cross-Linking, Ligation and Sequencing of Hybrids (CLASH)
4.3.4. Techniques for Investigating lncRNA-Protein Interaction
RNA Immunoprecipitation (RIP)
High-Throughput Sequencing of RNA Isolated by Cross-Linking Immunoprecipitation (HITS-CLIP)
Photoactivatable Ribonucleotide-Enhanced Cross Linking and Immunoprecipitation (PAR-CLIP)
5. Functional Involvement of lncRNAs in Endothelial Dysfunction-Associated Diseases
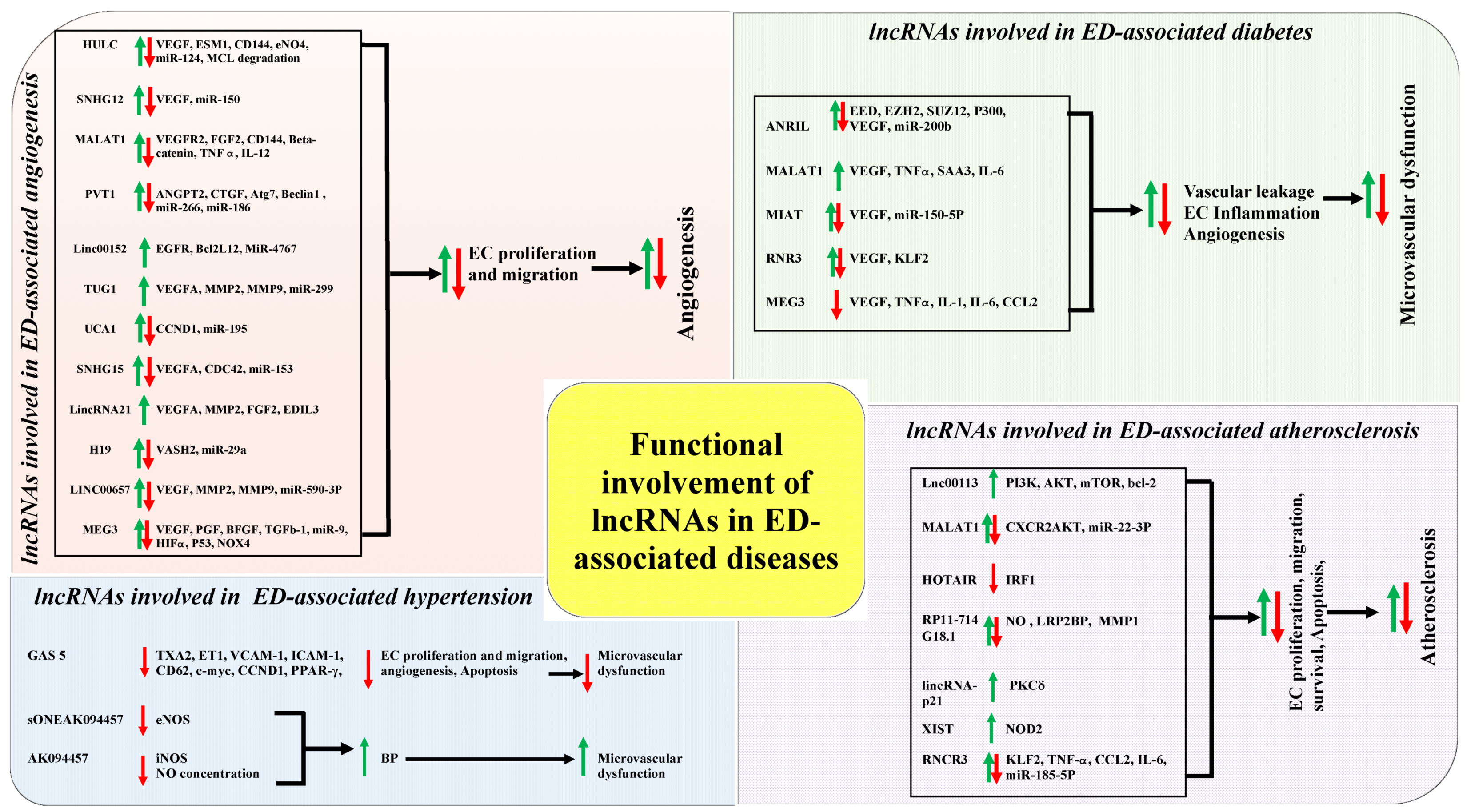
5.1. The Correlations of Functional Involvement of lncRNAs in ED and Angiogenesis
5.2. The Correlations of Functional Involvement of lncRNAs in ED and Diabetes
5.3. The Correlations of Functional Involvement of lncRNAs in ED and Hypertension
5.4. The Correlations of Functional Involvement of lncRNAs in ED and Atherosclerosis
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 6SG | 6-Thioguanosine |
| 4SU | 4-Thiouridine |
| AAs | Amino acids |
| ADAR | Adenosine deaminase acting on RNA |
| Akt | AKT serine/threonine kinase 1 |
| Alu | Arthrobacter luteus |
| AMT | Aminomethyltrioxalen |
| ANGPT2 | Angiopoietin 2 |
| ASOs | Antisense oligonucleotides |
| BACE1 | Beta-secretase 1 |
| BACE1AS | BACE1 (beta-secretase 1)-antisense RNA |
| BCYRN1 | Brain cytoplasmic RNA 1 |
| BP | Blood pressure |
| CAGE | Cap analysis of gene expression |
| CCNA2 | Cyclin A2 |
| CCNB1 | Cyclin B1 |
| CCNB2 | Cyclin B2 |
| Cdc42 | Cell division cycle 42 |
| CeRNA | Competing endogenous RNA |
| cGMP | Cyclic guanosine 3′, 5′-monophosphate |
| CHART | Capture hybridization analysis of RNA target |
| ChIRP | Chromatin isolation by RNA purification |
| CLASH | Cross-linking, ligation and sequencing of hybrids |
| COUP-TFII | Chicken ovalbumin upstream promoter transcription-factor-2 |
| CRISPRi | CRISPR interference |
| CTD | Carboxyl-teminal domain |
| CTGF | Connective tissue growth factor |
| CVD | Cardiovascular disease |
| CX3CL1 | C-X3-C motif chemokine ligand 1 |
| DSC | Disuccinimidyl glutarate |
| EC | Endothelial cell |
| ED | Endothelial dysfunction |
| EDHF | Endothelium derived hyperpolarizing factor |
| EeD | Embryonic ectoderm development |
| ERK | Extracellular signal-regulated kinase |
| ESM-1 | Endothelial cell specific molecule |
| EPC | Endothelial progenitor cell |
| EPC2 | Enhancer of polycomb homolog 2 |
| ER | Endoplasmic reticulum |
| ET-1 | Endothelin-1 |
| FA | Formaldehyde |
| FAK | Focal adhesion kinase |
| FISH | Fluorescence in situ hybridization |
| FGF2 | Fibroblast Growth Factor 2 |
| GAS5 | Growth arrest specific 5 |
| GATA-AS | Antisense transcript of GATA6 |
| GRP78 | Glucose regulated protein 78 |
| GR | Glucocorticoid receptor |
| GTP | Guanosine triphosphate |
| H3K4me2 | Demethylation of lysine 4 residue on histone 3 |
| H3K4me3 | Trimethylation of lysine 4 of histone H3 |
| H3K27me3 | Histone 3 lysine 27 trimethylation |
| H19 | H19 imprinted maternally expressed transcript |
| HDR | Homology direct repair |
| HITS-CLIP | High-throughput sequencing of RNA isolated by cross-linking immunoprecipitation |
| HOTAIR | HOX transcript antisense RNA |
| HULC | Highly upregulated in liver cancer |
| IL-6 | Interleukin-6 |
| HUVECs | Human umbilical vein endothelial cells |
| IL-1β | Interleukin 1 beta |
| KLF2 | Kruppel like factor |
| KRAB | Kruppel-associated box |
| LDL | Low-density lipoprotein |
| lncRNAs | Long non-coding RNAs |
| LOX2 | Lysyl oxidase-like 2 |
| LRP2BP | LRP2 binding protein |
| LSD1 | Lys-specific demethylase 1 |
| MALAT1 | Metastasis associated lung adenocarcinoma transcript 1 |
| MAPK | Mitogen-activated protein kinases |
| MCP-1 | Monocyte chemoattractant protein-1 |
| MEG3 | Maternally expressed 3 |
| MERFISH | Multiplexed error-robust fluorescence in situ hybridization |
| Mhrt | Myosin heavy chain associated RNA transcript |
| MIAT | Myocardial infarction associated transcript |
| MMP | Matrix metalloproteinase |
| MSC | Mesenchymal stem cell |
| mTOR | Mammalian target of rapamycin |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NATs | Natural antisense transcripts |
| NFAT | Nuclear factor of activated T-cells |
| NFkB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NGS | Next generation sequencing |
| NHEJ | Non-homologous end joining |
| NO | Nitric oxide |
| NOD2 | Nucleotide binding oligomerization domain containing 2 |
| eNOS | NO synthase |
| NR1 | Notoginsenoside R1 |
| NRON | Non-coding repressor of NFAT |
| NOX4 | NADPH Oxidase 4 |
| OGD | Oxygen-glucose deprivation |
| ORF | Open reading frame |
| oxLDL | Oxidized low-density lipoprotein |
| PAR-CLIP | Photoactivatable ribonucleotide-enhanced cross linking and immunoprecipitation |
| PGI2 | Prostacyclin |
| PGH2 | ProstaglandinH2 |
| PI3K | Phosphatidylinositol 3-kinase |
| PKCδ | Protein kinase C delta |
| PolII | RNA polymerase II |
| PRC2 | Polycomb repressive complex |
| PVT1 | Plasmacytoma variant translocation 1 |
| RAP | RNA antisense purification |
| RAP-RNA | RNA antisense purification followed by RNA sequencing |
| RBBP4 | RB binding protein 4, chromatin remodeling factor |
| RIP | RNA immunoprecipitation |
| RNA-FISH | RNA fluorescence in situ hybridization |
| RNA-Seq | RNA sequencing |
| RNAi | RNA interference |
| RNCR3 | Retinal non-coding RNA3 |
| ROS | Reactive oxygen species |
| SAGE | Serial analysis of gene expression |
| SAA3 | Serum amyloid antigen3 |
| sGC | Soluble guanylyl cyclase |
| sgRNA | Single guide RNA |
| shRNA | Short hairpin RNA |
| SINE | Short interspersed elements |
| siRNA | Small interfering RNA |
| SMAD6 | Mothers against decapentaplegic homologue 6 |
| smRNA-FISH | Single molecule RNA in situ hybridization |
| SMMECs | Skeletal muscle microvascular endothelial cells |
| SNHG12 | Small nucleolar RNA host gene 12 |
| SNHG15 | Small nucleolar RNA host gene 15 |
| SOX18 | SRY (Sex Determining Region Y)-Box 18 |
| SRY | Sex Determining Region Y |
| Suz12 | SUZ12 polycomb repressive complex 2 subunit |
| T1DM | Type 1 Diabetic Mellitus |
| T2DM | Type 2 Diabetic Mellitus |
| TAMs | Tumor-associated macrophages |
| TSLP | Thymic stromal lymphopoietin |
| TNF-α | Tumor necrosis factor-α |
| TSS | Transcriptional start site |
| TUG1 | Taurine up-regulated 1 |
| TXA2 | Thromboxane A2 |
| UBE2CP3 | Ubiquitin conjugating enzyme E2 C pseudogene 3 |
| UCA-1 | Urothelial cancer associated 1 |
| VCAM-1 | Vascular cell adhesion molecule-1 |
| VEC | vascular endothelial cells |
| VSMC | Vascular smooth muscle cells |
| VECs | Vascular endothelial cells |
| VEGF | Vascular endothelial growth factor |
| VEGFA | Vascular endothelial growth factor A |
| VEGFR2 | VEGF receptor 2 |
| XIST | X-inactive specific transcript. |
References
- Chlopicki, S. Perspectives in pharmacology of endothelium: From bench to bedside. Pharmacol. Rep. PR 2015, 67, vi–ix. [Google Scholar] [CrossRef] [PubMed]
- Lerman, A.; Zeiher, A.M. Endothelial function: Cardiac events. Circulation 2005, 1111, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Khazaei, M.; Moien-Afshari, F.; Laher, I. Vascular endothelial function in health and diseases. Pathophysiology 2008, 15, 49–67. [Google Scholar] [CrossRef] [PubMed]
- Godo, S.; Shimokawa, H. Endothelial functions. Arterioscler. Thromb. Vasc. Biol. 2017, 37, e108–e114. [Google Scholar] [CrossRef] [PubMed]
- Deanfield, J.E.; Halcox, J.P.; Rabelink, T.J. Endothelial function and dysfunction: Testing and clinical relevance. Circulation 2007, 115, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Nafisa, A.; Gray, S.G.; Cao, Y.; Wang, T.; Xu, S.; Wattoo, F.H.; Barras, M.; Cohen, N.; Kamato, D.; Little, P.J. Endothelial function and dysfunction: Impact of metformin. Pharmacol. Ther. 2018, 192, 150–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, A.; Gao, L.; Thakur, A.; Siu, P.M.; Lai, C.W.K. Role of free fatty acids in endothelial dysfunction. J. Biomed. Sci. 2017, 24, 50. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, B.; Economides, C.; Mayeda, G.; Burstein, S.; Kloner, R. The endothelial cell in health and disease: Its function, dysfunction, measurement and therapy. Int. J. Impot. Res. 2010, 22, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.M.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [Green Version]
- Nagano, T.; Fraser, P. No-nonsense functions for long noncoding RNAs. Cell 2011, 145, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Christopher, A.F.; Kaur, R.P.; Kaur, G.; Kaur, A.; Gupta, V.; Bansal, P. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect. Clin. Res. 2016, 7, 68–74. [Google Scholar] [PubMed]
- Selleri, L.; Bartolomei, M.S.; Bickmore, W.A.; He, L.; Stubbs, L.; Reik, W.; Barsh, G.S. A Hox-embedded long noncoding RNA: Is it all hot air? PLoS Genet. 2016, 12, e1006485. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Luo, S.; Peng, G.; Lu, J.Y.; Cui, G.; Liu, L.; Yan, P.; Yin, Y.; Liu, W.; Wang, R.; et al. Mouse knockout models reveal largely dispensable but context-dependent functions of lncRNAs during development. J. Mol. Cell Biol. 2018, 10, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, M.; Goff, L.A.; Lodato, S.; Bonev, B.; Groff, A.F.; Gerhardinger, C.; Sanchez-Gomez, D.B.; Hacisuleyman, E.; Li, E.; Spence, M.; et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2013, 2, e01749. [Google Scholar] [CrossRef] [Green Version]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Laurent, G.S.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015, 16, 20. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar] [CrossRef] [Green Version]
- Batista, P.J.; Chang, H.Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013, 152, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Wang, S. Angio-LncRs: LncRNAs that regulate angiogenesis and vascular disease. Theranostics 2018, 8, 3654–3675. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-K.; Hemberg, M.; Gray, J.M. Enhancer RNAs: A class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb. Perspect. Biol. 2015, 7, a018622. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Rosenfeld, M.G. Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 2016, 17, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Neumann, P.; Jaé, N.; Knau, A.; Glaser, S.F.; Fouani, Y.; Rossbach, O.; Krüger, M.; John, D.; Bindereif, A.; Grote, P.; et al. The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat. Commun. 2018, 9, 237. [Google Scholar] [CrossRef]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Morales, D.R.; Thomas, K.; Presser, A.; Bernstein, B.E.; Van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [Green Version]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Dykes, I.M.; Emanueli, C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef]
- Leisegang, M.S.; Fork, C.; Josipovic, I.; Richter, F.M.; Preussner, J.; Hu, J.; Miller, M.J.; Epah, J.; Hofmann, P.; Günther, S.; et al. Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation 2017, 136, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Bumgarner, S.L.; Neuert, G.; Voight, B.F.; Symbor-Nagrabska, A.; Grisafi, P.; Van Oudenaarden, A.; Fink, G.R. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol. Cell 2012, 45, 470–482. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Miyoshi, T.; Kugou, K.; Hoffman, C.S.; Shibata, T.; Ohta, K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 2008, 456, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Mariner, P.D.; Walters, R.D.; Espinoza, C.A.; Drullinger, L.F.; Wagner, S.D.; Kugel, J.F.; Goodrich, J.A. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell 2008, 29, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Martianov, I.; Ramadass, A.; Barros, A.S.; Chow, N.; Akoulitchev, A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007, 445, 666–670. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest–and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef] [PubMed]
- Willingham, A.T.; Orth, A.P.; Batalov, S.; Peters, E.C.; Wen, B.G.; Aza-Blanc, P.; Hogenesch, J.B.; Schultz, P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005, 309, 1570–1573. [Google Scholar] [CrossRef]
- Moreau, P.R.; Örd, T.; Downes, N.L.; Niskanen, H.; Bouvy-Liivrand, M.; Aavik, E.; Ylä-Herttuala, S.; Kaikkonen, M.U. Transcriptional profiling of hypoxia-regulated non-coding RNAs in human primary endothelial cells. Front. Cardiovasc. Med. 2018, 5, 159. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009, 10, 637–643. [Google Scholar] [CrossRef]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; Laurent, G.S.; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Faghihi, M.A.; Zhang, M.; Huang, J.; Modarresi, F.; Van Der Brug, M.P.; A Nalls, M.; Cookson, M.R.; St-Laurent, G.; Wahlestedt, C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010, 11, R56. [Google Scholar] [CrossRef] [PubMed]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Robb, G.B.; Carson, A.R.; Tai, S.C.; Fish, J.E.; Singh, S.; Yamada, T.; Scherer, S.W.; Nakabayashi, K.; Marsden, P.A.; Scherer, S.; et al. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J. Biol. Chem. 2004, 279, 37982–37996. [Google Scholar] [CrossRef] [PubMed]
- Fish, J.E.; Matouk, C.C.; Yeboah, E.; Bevan, S.C.; Khan, M.; Patil, K.; Ohh, M.; Marsden, P.A. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J. Biol. Chem. 2007, 282, 15652–15666. [Google Scholar] [CrossRef] [PubMed]
- Libby, P. Mechanisms of acute coronary syndromes and their implications for therapy. N. Engl. J. Med. 2013, 368, 2004–2013. [Google Scholar] [CrossRef]
- Libby, P. Inflammation in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2045–2051. [Google Scholar] [CrossRef]
- Mullick, A.E.; Soldau, K.; Kiosses, W.B.; Bell, T.A.; Tobias, P.S.; Curtiss, L.K. Increased endothelial expression of Toll-like receptor 2 at sites of disturbed blood flow exacerbates early atherogenic events. J. Exp. Med. 2008, 205, 373–383. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, X.; Qi, L.; Cai, Y.; Yang, P.; Xuan, G.; Jiang, Y. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget 2016, 7, 14429–14440. [Google Scholar] [CrossRef] [Green Version]
- Yin, D.; Li, Y.; Fu, C.; Feng, Y. Pro-Angiogenic Role of lncRNA HULC in Microvascular Endothelial Cells via Sequestrating miR-124. Cell. Physiol. Biochem. 2018, 50, 2188–2202. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, X.; Hamblin, M.H.; Yin, K.-J. Long Non-Coding RNA Malat1 Regulates Angiogenesis in Hindlimb Ischemia. Int. J. Mol. Sci. 2018, 19, 1723. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.K.; Ma, L.; Song, W.H.; Lu, B.Y.; Huang, Y.B.; Dong, H.M.; Ma, X.K.; Zhu, Z.Z.; Zhou, R. lncRNA-MALAT1 promotes angiogenesis of thyroid cancer by modulating tumor-associated macrophage FGF2 protein secretion. J. Cell. Biochem. 2017, 118, 4821–4830. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, Z.; Yuan, J.; Sun, L.; Lin, L.; Huang, N.; Bin, J.; Liao, Y.; Liao, W. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017, 395, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Tee, A.E.; Liu, B.; Song, R.; Li, J.; Pasquier, E.; Cheung, B.B.; Jiang, C.; Marshall, G.M.; Haber, M.; Norris, M.D.; et al. The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression. Oncotarget 2016, 7, 8663–8675. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-Y.; Yao, J.; Li, X.-M.; Song, Y.-C.; Wang, X.-Q.; Li, Y.-J.; Yan, B.; Jiang, Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014, 5, e1506. [Google Scholar] [CrossRef] [PubMed]
- Puthanveetil, P.; Chen, S.; Feng, B.; Gautam, A.; Chakrabarti, S. Long non-coding RNA MALAT 1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell. Mol. Med. 2015, 19, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Cheng, Y.G.; Huang, X.; Zhong, M.W.; Liu, S.Z.; Hu, S.Y. Downregulation of lncRNA MALAT1 contributes to renal functional improvement after duodenal-jejunal bypass in a diabetic rat model. J. Physiol. Biochem. 2018, 74, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Jin, X.; Xiang, Y.; Chen, Y.; Shen, C.-X.; Zhang, Y.-C.; Li, Y.-G. The lncRNA MALAT1 protects the endothelium against ox-LDL-induced dysfunction via upregulating the expression of the miR-22-3p target genes CXCR2 and AKT. FEBS Lett. 2015, 589, 3189–3196. [Google Scholar] [CrossRef] [Green Version]
- Michalik, K.M.; You, X.; Manavski, Y.; Doddaballapur, A.; Zörnig, M.; Braun, T.; John, D.; Ponomareva, Y.; Chen, W.; Uchida, S.; et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014, 114, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Xin, J.-W.; Jiang, Y.-G. Long noncoding RNA MALAT1 inhibits apoptosis induced by oxygen-glucose deprivation and reoxygenation in human brain microvascular endothelial cells. Exp. Ther. Med. 2017, 13, 1225–1234. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.Y.; Yu, M.S.; Li, X.; Zhang, Z.; Han, C.R.; Yan, B. Overexpression of long non-coding RNA MEG3 suppresses breast cancer cell proliferation, invasion, and angiogenesis through AKT pathway. Tumor Biol. 2017, 39, 1010428317701311. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Xie, W.; Shang, Q.; Su, B. The long noncoding RNA MEG3 is downregulated and inversely associated with VEGF levels in osteoarthritis. BioMed Res. Int. 2015, 2015, 356893. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Yang, W.; Yang, J.; Ding, J.; Li, S.; Wu, H.; Zhou, F.; Jiang, Y.; Teng, L.; Yang, J. Long noncoding RNA MEG3 negatively regulates proliferation and angiogenesis in vascular endothelial cells. DNA Cell Biol. 2017, 36, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Q.; Zhang, K.S.; Hu, B.; Niu, X.; Zhou, S.M.; Li, S.G.; Luo, Y.P.; Wang, Y.; Deng, Z.F. Downregulation of the long non-coding RNA Meg3 promotes angiogenesis after ischemic brain injury by activating notch signaling. Mol. Neurobiol. 2017, 54, 8179–8190. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Q.; Zhang, K.S.; Hu, B.; Niu, X.; Zhou, S.M.; Li, S.G.; Luo, Y.P.; Wang, Y.; Deng, Z.F. Long noncoding RNA MEG3 mediated angiogenesis after cerebral infarction through regulating p53/NOX4 axis. Biochem. Biophys. Res. Commun. 2017, 490, 700–706. [Google Scholar]
- Ruan, W.; Zhao, F.; Zhao, S.; Zhang, L.; Shi, L.; Pang, T. Knockdown of long noncoding RNA MEG3 impairs VEGF-stimulated endothelial sprouting angiogenesis via modulating VEGFR2 expression in human umbilical vein endothelial cells. Gene 2018, 649, 32–39. [Google Scholar] [CrossRef]
- Qiu, G.-Z.; Tian, W.; Fu, H.-T.; Li, C.-P.; Liu, B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem. Biophys. Res. Commun. 2016, 471, 135–141. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Di, L.L.; Zhu, J.; Sun, Y.X.; Wang, L.; Liu, L.N. Long non-coding RNA MEG3 mediates high glucose-induced endothelial cell dysfunction. Int. J. Clin. Exp. Pathol. 2018, 11, 1088–1100. [Google Scholar]
- Cai, H.; Liu, X.; Zheng, J.; Xue, Y.; Ma, J.; Li, Z.; Xi, Z.; Bao, M.; Liu, Y. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene 2017, 36, 318–331. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, T.; Zeng, X.; Li, G.; Du, L.; Ma, Z.; Wan, J.; Yang, Y. Effect of silencing lncRNATUG1 on rapamycin-induced inhibition of endothelial cell proliferation and migration. Exp. Ther. Med. 2018, 16, 1891–1899. [Google Scholar] [CrossRef]
- Dong, R.; Liu, G.-B.; Liu, B.-H.; Chen, G.; Li, K.; Zheng, S.; Dong, K.-R. Targeting long non-coding RNA-TUG1 inhibits tumor growth and angiogenesis in hepatoblastoma. Cell Death Dis. 2016, 7, e2278. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Hu, L.; Cheng, J.; Xu, J.; Zhong, Z.; Yang, Y.; Yuan, Z. lncRNA PVT1 promotes the angiogenesis of vascular endothelial cell by targeting miR-26b to activate CTGF/ANGPT2. Int. J. Mol. Med. 2018, 42, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, P.; Xue, Y.; Qu, C.; Zheng, J.; Liu, X.; Ma, J.; Liu, Y. PVT1 affects growth of glioma microvascular endothelial cells by negatively regulating miR-186. Tumor Biol. 2017, 39, 1010428317694326. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, D.; Ji, T.F.; Shi, L.; Yu, J.L. Overexpression of lncRNA ANRIL up-regulates VEGF expression and promotes angiogenesis of diabetes mellitus combined with cerebral infarction by activating NF-κB signaling pathway in a rat model. Oncotarget 2017, 8, 17347–17359. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.A.; Feng, B.; Chakrabarti, S. ANRIL: A regulator of VEGF in diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.M.; Lu, Y.F.; Hu, B.G.; Liang, W.C.; Zhu, X.; Yang, H.D.; Li, G.; Zhang, J.F. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget 2016, 7, 4712–4723. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Meng, K.; Jiang, L.; Zhong, Y.; Yang, Y.; Lan, Y.; Zeng, Q.; Cheng, L. Thymic stromal lymphopoietin-induced HOTAIR activation promotes endothelial cell proliferation and migration in atherosclerosis. Biosci. Rep. 2017, 37, BSR20170351. [Google Scholar] [CrossRef]
- Shan, K.; Jiang, Q.; Wang, X.-Q.; Wang, Y.-N.-Z.; Yang, H.; Yao, M.-D.; Liu, C.; Li, X.-M.; Yao, J.; Liu, B.; et al. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 2016, 7, e2248. [Google Scholar] [CrossRef]
- Shan, K.; Li, C.-P.; Liu, C.; Liu, X.; Yan, B. RNCR3: A regulator of diabetes mellitus-related retinal microvascular dysfunction. Biochem. Biophys. Res. Commun. 2017, 482, 777–783. [Google Scholar] [CrossRef]
- Xu, X.; Ma, C.; Liu, C.; Duan, Z.; Zhang, L. Knockdown of long noncoding RNA XIST alleviates oxidative low-density lipoprotein-mediated endothelial cells injury through modulation of miR-320/NOD2 axis. Biochem. Biophys. Res. Commun. 2018, 503, 586–592. [Google Scholar] [CrossRef]
- Jia, P.; Cai, H.; Liu, X.; Chen, J.; Ma, J.; Wang, P.; Liu, Y.; Zheng, J.; Xue, Y. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016, 381, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Weirick, T.; Militello, G.; Uchida, S. Long non-coding RNAs in endothelial biology. Front. Physiol. 2018, 9, 522. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, J.; Tang, N. Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience 2017, 354, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-Y.; Zhao, Z.-W.; Li, W.-M.; Yang, G.; Jing, P.-Y.; Li, P.; Dang, H.-Z.; Chen, Z.; Zhou, Y.-A.; Li, X.-F. Knockdown of MALAT1 expression inhibits HUVEC proliferation by upregulation of miR-320a and downregulation of FOXM1 expression. Oncotarget 2017, 8, 61499–61509. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashi, K.; Henderson, L.; Bonetti, A.; Carninci, P. Discovery and functional analysis of lncRNAs: Methodologies to investigate an uncharacterized transcriptome. Biochim. Biophys. Acta 2016, 1859, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martín, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [Green Version]
- Mockler, T.C.; Ecker, J.R. Applications of DNA tiling arrays for whole-genome analysis. Genomics 2005, 85, 1–15. [Google Scholar] [CrossRef]
- Yazaki, J.; Gregory, B.D.; Ecker, J.R. Mapping the genome landscape using tiling array technology. Curr. Opin. Plant Biol. 2007, 10, 534–542. [Google Scholar] [CrossRef] [Green Version]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef]
- Hu, M.; Polyak, K. Serial analysis of gene expression. Nat. Protoc. 2006, 1, 1743–1760. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Lassmann, T.; Murata, M.; Carninci, P. 5′ end–centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc. 2012, 7, 542–561. [Google Scholar] [CrossRef] [PubMed]
- Shiraki, T.; Kondo, S.; Katayama, S.; Waki, K.; Kasukawa, T.; Kawaji, H.; Kodzius, R.; Watahiki, A.; Nakamura, M.; Arakawa, T.; et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. USA 2003, 100, 15776–15781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurosawa, J.; Nishiyori, H.; Hayashizaki, Y. Deep cap analysis of gene expression. In PCR Protocols; Springer: Berlin/Heidelberg, Germany, 2011; pp. 147–163. [Google Scholar]
- Takahashi, H.; Kato, S.; Murata, M.; Carninci, P. CAGE (cap analysis of gene expression): A protocol for the detection of promoter and transcriptional networks. In Gene Regulatory Networks; Springer: Berlin/Heidelberg, Germany, 2012; pp. 181–200. [Google Scholar]
- Kawaji, H.; Lizio, M.; Itoh, M.; Kanamori-Katayama, M.; Kaiho, A.; Nishiyori-Sueki, H.; Shin, J.W.; Kojima-Ishiyama, M.; Kawano, M.; Murata, M.; et al. Comparison of CAGE and RNA-seq transcriptome profiling using clonally amplified and single-molecule next-generation sequencing. Genome Res. 2014, 24, 708–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Quinn, E.M.; Cormican, P.; Kenny, E.M.; Hill, M.; Anney, R.; Gill, M.; Corvin, A.P.; Morris, D.W. Development of strategies for SNP detection in RNA-seq data: Application to lymphoblastoid cell lines and evaluation using 1000 Genomes data. PLoS ONE 2013, 8, e58815. [Google Scholar] [CrossRef]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Laubinger, S.; Zeller, G.; Henz, S.R.; Sachsenberg, T.; Widmer, C.K.; Naouar, N.; Vuylsteke, M.; Schölkopf, B.; Rätsch, G.; Weigel, D. At-TAX: A whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol. 2008, 9, R112. [Google Scholar] [CrossRef]
- Gibb, E.A.; Vucic, E.A.; Enfield, K.S.S.; Stewart, G.L.; Lonergan, K.M.; Kennett, J.Y.; Becker-Santos, D.D.; Macaulay, C.E.; Lam, S.; Brown, C.J.; et al. Human cancer long non-coding RNA transcriptomes. PLoS ONE 2011, 6, e25915. [Google Scholar] [CrossRef]
- Kodzius, R.; Kojima, M.; Nishiyori, H.; Nakamura, M.; Fukuda, S.; Tagami, M.; Sasaki, D.; Imamura, K.; Kai, C.; Harbers, M.; et al. CAGE: Cap analysis of gene expression. Nat. Methods 2006, 3, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Hon, C.-C.; Ramilowski, J.A.; Harshbarger, J.; Bertin, N.; Rackham, O.J.L.; Gough, J.; Denisenko, E.; Schmeier, S.; Poulsen, T.M.; Severin, J.; et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 2017, 543, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Kinne, J.; Donham, B.; Jiang, F.; Ding, L.; Hassler, J.R.; Kaufman, R.J. Read-Split-Run: An improved bioinformatics pipeline for identification of genome-wide non-canonical spliced regions using RNA-Seq data. BMC Genom. 2016, 17, 503. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Holt, R.A.; Jones, S.J. The new paradigm of flow cell sequencing. Genome Res. 2008, 18, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Cloonan, N.; Forrest, A.R.R.; Kolle, G.; A Gardiner, B.B.; Faulkner, G.J.; Brown, M.K.; Taylor, D.F.; Steptoe, A.L.; Wani, S.; Bethel, G.; et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods 2008, 5, 613–619. [Google Scholar] [CrossRef]
- McFadden, E.J.; Hargrove, A.E. Biochemical methods to investigate lncRNA and the influence of lncRNA: Protein complexes on chromatin. Biochemistry 2016, 55, 1615–1630. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Chang, K.; Marran, K.; Valentine, A.; Hannon, G.J. RNAi in cultured mammalian cells using synthetic siRNAs. Cold Spring Harb. Protoc. 2012, 2012, 957–961. [Google Scholar] [CrossRef]
- Rao, D.D.; Vorhies, J.S.; Senzer, N.; Nemunaitis, J. siRNA vs. shRNA: Similarities and differences. Adv. Drug Deliv. Rev. 2009, 61, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Sledz, C.A.; Williams, B.R.G. RNA interference in biology and disease. Blood 2005, 106, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.Q.; Chang, D.C. The gene-silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochem. Biophys. Res. Commun. 2004, 318, 303–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.J.; Norrbom, M.; Chun, S.; Bennett, C.F.; Rigo, F. Nonsense-mediated decay as a terminating mechanism for antisense oligonucleotides. Nucleic Acids Res. 2014, 42, 5871–5879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, N.; Stein, C. Antisense oligonucleotides: Basic concepts and mechanisms. Mol. Cancer Ther. 2002, 1, 347–355. [Google Scholar] [PubMed]
- Lennox, K.A.; Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2015, 44, 863–877. [Google Scholar] [CrossRef] [Green Version]
- Ho, T.-T.; Zhou, N.; Huang, J.; Koirala, P.; Xu, M.; Fung, R.; Wu, F.; Mo, Y.-Y. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 2014, 43, e17. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Bassett, A.R.; Akhtar, A.; Barlow, D.P.; Bird, A.P.; Brockdorff, N.; Duboule, D.; Ephrussi, A.; Ferguson-Smith, A.C.; Gingeras, T.R.; Haerty, W.; et al. Science Forum: Considerations when investigating lncRNA function in vivo. eLife 2014, 3, e03058. [Google Scholar] [CrossRef]
- Liu, Z.; Hui, Y.; Shi, L.; Chen, Z.; Xu, X.; Chi, L.; Fan, B.; Fang, Y.; Liu, Y.; Ma, L.; et al. Efficient CRISPR/Cas9-mediated versatile, predictable, and donor-free gene knockout in human pluripotent stem cells. Stem Cell Rep. 2016, 7, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Han, X.; Yuan, J.; Geng, T.; Chen, S.; Hu, X.; Cui, I.H.; Cui, H. Biallelic Insertion of a Transcriptional Terminator via CRISPR/Cas9 Efficiently Silences Expression of Protein-coding and Non-coding RNA Genes. J. Biol. Chem. 2017, 292, 5624–5633. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Tibbit, C.; Liu, J.-L. Effective knockdown of Drosophila long non-coding RNAs by CRISPR interference. Nucleic Acids Res. 2016, 44, e84. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; Attenello, F.J.; Villalta, J.E.; Cho, M.Y.; Chen, Y.; et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017, 355, eaah7111. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watts, J.K.; Corey, D.R. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012, 226, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Stojic, L.; Niemczyk, M.; Orjalo, A.; Ito, Y.; Ruijter, A.E.M.; Uribe-Lewis, S.; Joseph, N.; Weston, S.; Menon, S.; Odom, D.T.; et al. Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat. Commun. 2016, 7, 10406. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.F.; Swayze, E.E. RNA targeting therapeutics: Molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 259–293. [Google Scholar] [CrossRef]
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012, 482, 331–338. [Google Scholar] [CrossRef]
- Han, J.; Zhang, J.; Chen, L.; Shen, B.; Zhou, J.; Hu, B.; Du, Y.; Tate, P.H.; Huang, X.; Zhang, W. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol. 2014, 11, 829–835. [Google Scholar] [CrossRef]
- Eißmann, M.; Gutschner, T.; Hämmerle, M.; Günther, S.; Caudron-Herger, M.; Groß, M.; Schirmacher, P.; Rippe, K.; Braun, T.; Diederichs, S.; et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012, 9, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.-F.; Yin, Q.-F.; Chen, T.; Zhang, Y.; Zhang, X.-O.; Wu, Z.; Zhang, S.; Wang, H.-B.; Ge, J.; Lu, X.; et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Wang, H.; Shivalila, C.S.; Cheng, A.W.; Shi, L.; Jaenisch, R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013, 154, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, A.A.; Lim, W.A.; Qi, L.S. Beyond editing: Repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016, 17, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimarães, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Cong, L.; Yan, W.X.; Ran, F.A.; Zetsche, B.; Li, Y.; Kurabayashi, A.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Staphylococcus aureus Cas9. Cell 2015, 162, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Ma, Q.; Tanasa, B.; Nunez, E.; Chen, A.Y.; Merkurjev, D.; Zhang, J.; Ohgi, K.; Song, X.; et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013, 498, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Carlevaro-Fita, J.; Johnson, R. Global positioning system: Understanding long noncoding RNAs through subcellular localization. Mol. Cell 2019, 73, 869–883. [Google Scholar] [CrossRef] [PubMed]
- Carlevaro-Fita, J.; Rahim, A.; Guigó, R.; Vardy, L.A.; Johnson, R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 2016, 22, 867–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashid, F.; Shah, A.; Shan, G. Long non-coding RNAs in the cytoplasm. Genom. Proteom. Bioinform. 2016, 14, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Hao, Q.; Prasanth, K.V. Nuclear long noncoding RNAs: Key regulators of gene expression. Trends Genet. 2018, 34, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Bogaard, P.V.D.; Rifkin, S.A.; Van Oudenaarden, A.; Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 2008, 5, 877–879. [Google Scholar] [CrossRef] [Green Version]
- Dunagin, M.; Cabili, M.N.; Rinn, J.; Raj, A. Visualization of lncRNA by single-molecule fluorescence in situ hybridization. In Nuclear Bodies and Noncoding RNAs; Springer: Berlin/Heidelberg, Germany, 2015; pp. 3–19. [Google Scholar]
- Vance, K.W. Mapping long noncoding RNA chromatin occupancy using capture hybridization analysis of RNA targets (CHART). In Enhancer RNAs; Springer: Berlin/Heidelberg, Germany, 2017; pp. 39–50. [Google Scholar]
- Sexton, A.N.; Machyna, M.; Simon, M.D. Capture hybridization analysis of DNA targets. In Polycomb Group Proteins; Springer: Berlin/Heidelberg, Germany, 2016; pp. 87–97. [Google Scholar]
- Chu, C.; Quinn, J.; Chang, H.Y. Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. JoVE 2012, 61, e3912. [Google Scholar] [CrossRef]
- Faoro, C.; Ataide, S.F. Ribonomic approaches to study the RNA-binding proteome. FEBS Lett. 2014, 588, 3649–3664. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.; Lander, E.S.; Guttman, M. RNA antisense purification (RAP) for mapping RNA interactions with chromatin. Methods Mol. Biol. (Clifton NJ) 2015, 1262, 183–197. [Google Scholar]
- Engreitz, J.M.; Pandya-Jones, A.; McDonel, P.; Shishkin, A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S.; et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013, 341, 1237973. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.M.; Sirokman, K.; McDonel, P.; Shishkin, A.A.; Surka, C.; Russell, P.; Grossman, S.R.; Chow, A.Y.; Guttman, M.; Lander, E.S. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 2014, 159, 188–199. [Google Scholar] [CrossRef]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Kudla, G.; Granneman, S.; Hahn, D.; Beggs, J.D.; Tollervey, D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 10010–10015. [Google Scholar] [CrossRef] [PubMed]
- Hausser, J.; Zavolan, M. Identification and consequences of miRNA-target interactions--beyond repression of gene expression. Nat. Rev. Genet. 2014, 15, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Zhang, Q.C.; Lee, B.; Flynn, R.A.; Smith, M.A.; Robinson, J.T.; Davidovich, C.; Gooding, A.R.; Goodrich, K.J.; Mattick, J.S.; et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 2016, 165, 1267–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gagliardi, M.; Matarazzo, M.R. RIP: RNA Immunoprecipitation. Methods Mol. Biol. (Clifton NJ) 2016, 1480, 73–86. [Google Scholar]
- Zhao, J.; Ohsumi, T.K.; Kung, J.T.; Ogawa, Y.; Grau, D.J.; Sarma, K.; Song, J.J.; Kingston, R.E.; Borowsky, M.; Lee, J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 2010, 40, 939–953. [Google Scholar] [CrossRef]
- Selth, L.A.; Close, P.; Svejstrup, J.Q. Studying RNA-protein interactions in vivo by RNA immunoprecipitation. Methods Mol. Biol. (Clifton NJ) 2011, 791, 253–264. [Google Scholar]
- Niranjanakumari, S.; Lasda, E.; Brazas, R.; Garcia-Blanco, M.A. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods (San Diego California) 2002, 26, 182–190. [Google Scholar] [CrossRef]
- Guil, S.; Soler, M.; Portela, A.; Carrère, J.; Fonalleras, E.; Gómez, A.; Villanueva, A.; Esteller, M.; Moruno, A.G. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat. Struct. Mol. Biol. 2012, 19, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Licatalosi, D.D.; Mele, A.; Fak, J.J.; Ule, J.; Kayikci, M.; Chi, S.W.; Clark, T.A.; Schweitzer, A.C.; Blume, J.E.; Wang, X.; et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 2008, 456, 464–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009, 460, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Darnell, R.B. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Biotechnol. 2011, 29, 607–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spitzer, J.; Hafner, M.; Landthaler, M.; Ascano, M.; Farazi, T.; Wardle, G.; Nusbaum, J.; Khorshid, M.; Burger, L.; Zavolan, M.; et al. PAR-CLIP (Photoactivatable Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation): A step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins. Methods Enzymol. 2014, 539, 113–161. [Google Scholar] [PubMed]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M.; Jungkamp, A.C.; Munschauer, M.; et al. PAR-CliP—A method to identify transcriptome-wide the binding sites of RNA binding proteins. J. Vis. Exp. JoVE 2010, 41, e2034. [Google Scholar] [CrossRef]
- Ascano, M.; Hafner, M.; Cekan, P.; Gerstberger, S.; Tuschl, T. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip. Rev. RNA 2012, 3, 159–177. [Google Scholar] [CrossRef]
- Fok, E.T.; Scholefield, J.; Fanucchi, S.; Mhlanga, M.M. The emerging molecular biology toolbox for the study of long noncoding RNA biology. Epigenomics 2017, 9, 1317–1327. [Google Scholar] [CrossRef]
- Shukla, C.J.; McCorkindale, A.L.; Gerhardinger, C.; Korthauer, K.D.; Cabili, M.N.; Shechner, D.M.; Irizarry, R.A.; Maass, P.G.; Rinn, J.L. High-throughput identification of RNA nuclear enrichment sequences. EMBO J. 2018, 37, e98452. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.-J.; Lee, J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Karamysheva, A. Mechanisms of angiogenesis. Biochemistry (Moscow) 2008, 73, 751. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P. Angiogenesis in health and disease. Nat. Med. 2003, 9, 653–660. [Google Scholar] [CrossRef] [PubMed]
- LaGory, E.L.; Giaccia, A.J. The ever-expanding role of HIF in tumour and stromal biology. Nat. Cell Biol. 2016, 18, 356–365. [Google Scholar] [CrossRef] [Green Version]
- Muz, B.; De La Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef]
- Cuadrado-Godia, E.; Regueiro, A.; Núñez, J.; Díaz-Ricard, M.; Novella, S.; Oliveras, A.; Valverde, M.A.; Marrugat, J.; Ois, Á.; Giralt-Steinhauer, E.; et al. Endothelial progenitor cells predict cardiovascular events after atherothrombotic stroke and acute myocardial infarction. A PROCELL Substudy. PLoS ONE 2015, 10, e0132415. [Google Scholar] [CrossRef]
- Ali, M.A.; Elsayed, A.S.; Ali, M.M.; Shafei, A.E.S.; Ghanem, H.G.; I Shehata, A.; Abdelgawad, A.A.; Handal, H.R.; Talaat, K.A.; Ashaal, A.E.; et al. Mechanistic effects of mesenchymal and hematopoietic stem cells: New therapeutic targets in myocardial infarction. J. Cell. Biochem. 2018, 119, 5274–5286. [Google Scholar]
- Ferrara, N.; Kerbel, R.S. Angiogenesis as a therapeutic target. Nature 2005, 438, 967–974. [Google Scholar] [CrossRef]
- M Kumar, M.; Goyal, R. lncRNA as a therapeutic target for angiogenesis. Curr. Top. Med. Chem. 2017, 17, 1750–1757. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, J.; Xi, X.; Tan, N.; Zhang, L. SNHG12 Promotes Angiogenesis Following Ischemic Stroke via Regulating miR-150/VEGF Pathway. Neuroscience 2018, 390, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Cao, S.; Wang, Y.; Hu, Y.; Liu, H.; Li, J.; Chen, J.; Li, P.; Liu, J.; Wang, Q.; et al. Long non-coding RNA UBE2CP3 enhances HCC cell secretion of VEGFA and promotes angiogenesis by activating ERK1/2/HIF-1α/VEGFA signalling in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 113. [Google Scholar] [CrossRef] [PubMed]
- Teng, W.; Qiu, C.; He, Z.; Wang, G.; Xue, Y.; Hui, X. Linc00152 suppresses apoptosis and promotes migration by sponging miR-4767 in vascular endothelial cells. Oncotarget 2017, 8, 85014–85023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, D.; Fu, C.; Sun, D. Silence of lncRNA UCA1 Represses the Growth and Tube Formation of Human Microvascular Endothelial Cells Through miR-195. Cell. Physiol. Biochem. 2018, 49, 1499–1511. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, Y.; Li, Z.; Zheng, S.; Wang, Z.; Li, W.; Bi, Z.; Li, L.; Jiang, Y.; Luo, Y.; et al. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J. Cell. Mol. Med. 2018, 22, 655–667. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, Y.; Liu, X.; Qu, C.; Cai, H.; Wang, P.; Li, Z. SNHG15 affects the growth of glioma microvascular endothelial cells by negatively regulating miR-153. Oncol. Rep. 2017, 38, 3265–3277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellano, J.J.; Navarro, A.; Viñolas, N.; Marrades, R.M.; Moises, J.; Cordeiro, A.; Saco, A.; Muñoz, C.; Fuster, D.; Molins, L.; et al. LincRNA-p21 Impacts Prognosis in Resected Non-Small Cell Lung Cancer Patients through Angiogenesis Regulation. J. Thorac. Oncol. 2016, 11, 2173–2182. [Google Scholar] [CrossRef]
- Bao, M.H.; Li, G.Y.; Huang, X.S.; Tang, L.; Dong, L.P.; Li, J.M. Long non-coding RNA LINC00657 acting as miR-590-3p sponge to facilitate low concentration oxidized low-density lipoprotein-induced angiogenesis. Mol. Pharmacol. 2018, 93, 368–375. [Google Scholar] [CrossRef]
- Young, T.; Matsuda, T.; Cepko, C. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 2005, 15, 501–512. [Google Scholar] [CrossRef]
- Czauderna, P.; Lopez-Terrada, D.; Hiyama, E.; Häberle, B.; Malogolowkin, M.H.; Meyers, R.L. Hepatoblastoma state of the art: Pathology, genetics, risk stratification, and chemotherapy. Curr. Opin. Pediatr. 2014, 26, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Jia, D.; Xue, P.; Cui, X.; Li, K.; Zheng, S.; He, X.; Dong, K. Genome-wide analysis of long noncoding RNA (lncRNA) expression in hepatoblastoma tissues. PLoS ONE 2014, 9, e85599. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 non-coding RNA: A tumor suppressor. J. Mol. Endocrinol. 2012, 48, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ley, S.H.; Hu, F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol. 2018, 14, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Mellbin, L.G.; Anselmino, M.; Lars, R. Diabetes, prediabetes and cardiovascular risk. Eur. J. Cardiovasc. Prev. Rehabil. 2010, 17, s9–s14. [Google Scholar] [CrossRef] [PubMed]
- Low Wang, C.C.; Hess, C.N.; Hiatt, W.R.; Goldfine, A.B. Clinical update: Cardiovascular disease in diabetes mellitus: Atherosclerotic cardiovascular disease and heart failure in type 2 diabetes mellitus-mechanisms, management, and clinical considerations. Circulation 2016, 133, 2459–2502. [Google Scholar] [CrossRef]
- Roberts, A.C.; Porter, K.E. Cellular and molecular mechanisms of endothelial dysfunction in diabetes. Diabetes Vasc. Dis. Res. 2013, 10, 472–482. [Google Scholar] [CrossRef] [Green Version]
- Hamdy, O.; Ledbury, S.; Mullooly, C.; Jarema, C.; Porter, S.; Ovalle, K.; Moussa, A.; Caselli, A.; Caballero, A.E.; Economides, P.A.; et al. Lifestyle modification improves endothelial function in obese subjects with the insulin resistance syndrome. Diabetes Care 2003, 26, 2119–2125. [Google Scholar] [CrossRef]
- Hamburg, N.M.; Larson, M.G.; Vita, J.A.; Vasan, R.S.; Keyes, M.J.; Widlansky, M.E.; Fox, C.S.; Mitchell, G.F.; Levy, D.; Meigs, J.B.; et al. Metabolic syndrome, insulin resistance, and brachial artery vasodilator function in Framingham Offspring participants without clinical evidence of cardiovascular disease. Am. J. Cardiol. 2008, 101, 82–88. [Google Scholar] [CrossRef]
- Avogaro, A.; De Kreutzenberg, S.V.; Fadini, G.P. Endothelial dysfunction: Causes and consequences in patients with diabetes mellitus. Diabetes Res. Clin. Pract. 2008, 82, S94–S101. [Google Scholar] [CrossRef]
- Soldatos, G.; Cooper, M.E.; Jandeleit-Dahm, K.A.M. Advanced-glycation end products in insulin-resistant states. Curr. Hypertens. Rep. 2005, 7, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Tabit, C.E.; Chung, W.B.; Hamburg, N.M.; Vita, J.A. Endothelial dysfunction in diabetes mellitus: Molecular mechanisms and clinical implications. Rev. Endocr. Metab. Disord. 2010, 11, 61–74. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Ou, C.; Xiao, Y.; Han, Q.; Li, H.; Zhou, S. lncRNAs: Key players and novel insights into diabetes mellitus. Oncotarget 2017, 8, 71325–71341. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Mantella, L.-E.; Pan, Y.; Quan, A.; Sabongui, S.; Sandhu, P.; Teoh, H.; Al-Omran, M.; Verma, S. A global profile of glucose-sensitive endothelial-expressed long non-coding RNAs. Can. J. Physiol. Pharmacol. 2016, 94, 1007–1014. [Google Scholar] [CrossRef]
- Yan, B.; Yao, J.; Liu, J.Y.; Li, X.M.; Wang, X.Q.; Li, Y.J.; Tao, Z.F.; Song, Y.C.; Chen, Q.; Jiang, Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 2015, 116, 1143–1156. [Google Scholar] [CrossRef]
- Widder, J.D.; Fraccarollo, D.; Galuppo, P.; Hansen, J.M.; Jones, D.P.; Ertl, G.; Bauersachs, J. Attenuation of angiotensin II–induced vascular dysfunction and hypertension by overexpression of thioredoxin 2. Hypertension 2009, 54, 338–344. [Google Scholar] [CrossRef]
- Doughan, A.K.; Harrison, D.G.; Dikalov, S.I. Molecular mechanisms of angiotensin II–mediated mitochondrial dysfunction: Linking mitochondrial oxidative damage and vascular endothelial dysfunction. Circ. Res. 2008, 102, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Vecchione, C.; Carnevale, D.; Di Pardo, A.; Gentile, M.T.; Damato, A.; Cocozza, G.; Antenucci, G.; Mascio, G.; Bettarini, U.; Landolfi, A.; et al. Pressure-induced vascular oxidative stress is mediated through activation of integrin-linked kinase 1/βPIX/Rac-1 pathway. Hypertension 2009, 54, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Magen, E.; Feldman, A.; Mishal, J.; Cohen, Z.; Schlezinger, M.; Ben Alon, D.; Linov, L. Potential link between C3a, C3b and endothelial progenitor cells in resistant hypertension. Am. J. Med. Sci. 2010, 339, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.M.; Zalos, G.; Halcox, J.P.J.; Schenke, W.H.; Waclawiw, M.A.; Quyyumi, A.A.; Finkel, T. Circulating endothelial progenitor cells, vascular function, and cardiovascular risk. N. Engl. J. Med. 2003, 348, 593–600. [Google Scholar] [CrossRef]
- Hage, F.G.; Oparil, S.; Xing, D.; Chen, Y.-F.; McCrory, M.A.; Szalai, A.J. C-reactive protein-mediated vascular injury requires complement. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1189–1195. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Xie, H.; Zhan, H.; Li, J.; Liu, Y.; Huang, W. Prognostic values of long noncoding RNA GAS5 in various carcinomas: An updated systematic review and meta-analysis. Front. Physiol. 2017, 8, 814. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.T.; Mourtada-Maarabouni, M.; Farzaneh, F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem. Soc. Trans. 2011, 39, 482–486. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.Z.; Shan, K.; Yao, M.D.; Yao, J.; Wang, J.J.; Li, X.; Liu, B.; Zhang, Y.Y.; Ji, Y.; Jiang, Q.; et al. Long noncoding RNA-GAS5: A novel regulator of hypertension-induced vascular remodeling. Hypertension 2016, 68, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, X.; Lin, Y.; Suo, M.; Gong, L.; Chen, J.; Hui, R. Anti-hypertensive effect of Lycium barbarum L. with down-regulated expression of renal endothelial lncRNA sONE in a rat model of salt-sensitive hypertension. Int. J. Clin. Exp. Pathol. 2015, 8, 6981–6987. [Google Scholar] [PubMed]
- Yang, Y.; Xi, P.; Xie, Y.; Zhao, C.; Xu, J.; Jiang, J. Notoginsenoside R1 reduces blood pressure in spontaneously hypertensive rats through a long non-coding RNA AK094457. Int. J. Clin. Exp. Pathol. 2015, 8, 2700–2709. [Google Scholar] [PubMed]
- Hansson, G.K. Inflammation, atherosclerosis, and coronary artery disease. N. Engl. J. Med. 2005, 352, 1685–1695. [Google Scholar] [CrossRef] [PubMed]
- Rafieian-Kopaei, M.; Setorki, M.; Doudi, M.; Baradaran, A.; Nasri, H. Atherosclerosis: Process, indicators, risk factors and new hopes. Int. J. Prev. Med. 2014, 5, 927–946. [Google Scholar] [PubMed]
- Tabas, I.; García-Cardeña, G.; Owens, G.K. Recent insights into the cellular biology of atherosclerosis. J. Cell Biol. 2015, 209, 13–22. [Google Scholar] [CrossRef]
- Park, K.-H.; Park, W.J. Endothelial dysfunction: Clinical implications in cardiovascular disease and therapeutic approaches. J. Korean Med. Sci. 2015, 30, 1213–1225. [Google Scholar] [CrossRef]
- Hanson, M.; Gluckman, P. Endothelial dysfunction and cardiovascular disease: The role of predictive adaptive responses. Heart 2005, 91, 864–866. [Google Scholar] [CrossRef] [PubMed]
- Pober, J.S.; Sessa, W.C. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 2007, 7, 803–815. [Google Scholar] [CrossRef] [PubMed]
- Aryal, B.; Suárez, Y. Non-coding RNA regulation of endothelial and macrophage functions during atherosclerosis. Vasc. Pharmacol. 2018, 114, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Qian, M.; Kyler, K.; Xu, J. Endothelial-Vascular Smooth Muscle Cells Interactions in Atherosclerosis. Front. Cardiovasc. Med. 2018, 5, 151. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, H.; Ge, J. Long noncoding RNA: Recent updates in atherosclerosis. Int. J. Biol. Sci. 2016, 12, 898–910. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Yan, C.; Zhang, L.; Li, Y.; Wan, Q. lncRNA ENST00113 promotes proliferation, survival, and migration by activating PI3K/Akt/mTOR signaling pathway in atherosclerosis. Medicine 2018, 97, e0473. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, L.; Xu, B.M.; Tang, W.H.; Ye, Z.D.; Huang, C.; Ma, X.; Zhao, J.J.; Guo, F.X.; Kang, C.M.; et al. Lnc RNA-RP 11-714G18. 1 suppresses vascular cell migration via directly targeting LRP 2 BP. Immunol. Cell Biol. 2018, 96, 175–189. [Google Scholar] [CrossRef]
- Zhou, T.; Chen, X. Long intergenic noncoding RNA p21 mediates oxidized LDL-induced apoptosis and expression of LOX-1 in human coronary artery endothelial cells. Mol. Med. Rep. 2017, 16, 8513–8519. [Google Scholar] [CrossRef]
- Halimulati, M.; Duman, B.; Nijiat, J.; Aizezi, A. Long noncoding RNA TCONS_00024652 regulates vascular endothelial cell proliferation and angiogenesis via microRNA-21. Exp. Ther. Med. 2018, 16, 3309–3316. [Google Scholar] [CrossRef]
- Liao, J.; He, Q.; Li, M.; Chen, Y.; Liu, Y.; Wang, J. lncRNA MIAT: Myocardial infarction associated and more. Gene 2016, 578, 158–161. [Google Scholar] [CrossRef]
- Han, P.; Li, W.; Lin, C.-H.; Yang, J.; Shang, C.; Nuernberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.-Y.; Lin, C.-J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.-Y.; Zhang, L.-X.; Tian, C.-J.; Tang, X.-Y.; Zhao, L.-M.; Guo, Y.-L.; Cheng, D.-J.; Chen, X.-L.; Ma, L.-J.; Chen, Z.-C. lncRNAs BCYRN1 promoted the proliferation and migration of rat airway smooth muscle cells in asthma via upregulating the expression of transient receptor potential 1. Am. J. Transl. Res. 2016, 8, 3409–3418. [Google Scholar] [PubMed]


| Method | Advantages | Disadvantages | References |
|---|---|---|---|
| Tiling arrays |
|
| [86,88,89] |
| SAGE |
|
| [90,91] |
| CAGE |
|
| [92,93,94,95,96] |
| RNA-Seq |
|
| [97,98,99] |
| Method | Advantages | Disadvantages | References |
|---|---|---|---|
| RNAi |
|
| [112,113,114,115] |
| ASOs |
|
| [116,117,118,119] |
| CRISPR/Cas9 |
|
| [120,121,122,123,124] |
| CRISPRi |
|
| [125,126] |
| Purpose of Study | Method | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Identification of subcellular localization | smRNA-FISH |
|
| [149,150] |
| Investigation of lncRNA-DNA interaction | CHART |
|
| [86,151,152] |
| ChIRP |
|
| [153,154,155] | |
| RAP |
|
| [156,157] | |
| Investigation of lncRNA-RNA interaction | RAP-RNA |
|
| [158] |
| CLASH |
|
| [159,160,161,162] | |
| Investigation of lncRNA-protein interaction | RIP |
|
| [163,164,165,166] |
| HITS-CLIP |
|
| [167,168,169,170] | |
| PAR-CLIP |
|
| [171,172,173] |
| Name | Location | Disease Association | Correlations of the Functional Involvement of lncRNAs in ED and Angiogenesis | References |
|---|---|---|---|---|
| HULC | 6p24.3 | Gliomas | Promotes angiogenesis through regulating endothelial cell specific molecule 1 (ESM-1) via PI3K/AKT/mTOR pathway | [49] |
| Angiogenesis | Regulates EC angiogenesis via sequestrating miR-124 | [50] | ||
| Small nucleolar RNA host gene 12 (SNHG12) | 1p35.3 | Ischemic stroke | Promotes angiogenesis of oxygen-glucose deprivation (OGD)-treated brain microvascular EC through regulating miR-150/VEGF pathway | [186] |
| MALAT1 | 11q13.1 | Hindlimb ischemia | Promotes cell-autonomous angiogenesis of skeletal muscle microvascular endothelial cells (SMMECs) via VEGFR2 regulation | [51] |
| Thyroid cancer | Promotes angiogenesis through regulating fibroblast Growth Factor 2 (FGF2) protein secretion from tumor-associated macrophages (TMAs) | [52] | ||
| Gastric cancer | Correlated with endothelial vessel density and promotes angiogenesis by regulating VE-cadherin/β-catenin complex, extracellular signal-regulated kinase (ERK)/ matrix metalloproteinase (MMP), and focal adhesion kinase (FAK)/paxillin signalizing pathways | [53] | ||
| Neuroblastoma | Promotes hypoxia-induced angiogenesis by increasing FGF2 expression | [54] | ||
| Ubiquitin conjugating enzyme E2 C pseudogene 3 (UBE2CP3) | 4q12 | Hepatocellular carcinoma | Stimulates vascular endothelial growth factor A (VEGFA) secretion and promotes HUVEC proliferation, migration and tube formation through activating ERK1/2/ hypoxia inducible factor 1 subunit alpha (HIF-1α)/VEGFA signalling pathways | [187] |
| PVT1 | 8q24.21 | Angiogenesis | Regulates connective tissue growth factor (CTGF) and angiopoietin 2 (ANGPT2) expression through interacting with miR-26b and promotes the angiogenesis of HUVEC | [72] |
| Glioma | Promotes glioma VEC proliferation, migration and angiogenesis by regulating miR-186 | [73] | ||
| Linc00152 | 2p11.2 | Angiogenesis | Promotes oxidized low-density lipoprotein (oxLDL)-treated HUVEC migration and inhibits apoptosis by sponging miR-4767 that regulates angiogenesis | [188] |
| TUG1 | 22q12.2 | Glioblastoma | Promotes glioblastoma-induced EC proliferation, migration and angiogenesis by inhibiting miR-299 | [69] |
| Angiogenesis | Regulates rapamycin-mediated inhibition of EC proliferation, migration and tube formation | [70] | ||
| Hepatoblastoma | Promotes angiogenesis via regulating VEGFA expression by sponging miR-34a | [71] | ||
| Urothelial cancer associated 1 (UCA1) | 19p13.12 | Angiogenesis | Promotes human microvascular endothelial cells (HMEC) proliferation, migration and tube formation through inhibiting miR-195 and activating mitogen-activated protein kinase kinase (MEK)/ERK/mTOR pathways | [189] |
| MEG3 | 14q32.2 | Breast cancer | Overexpressed MEG3 suppresses tumour growth and angiogenesis via inhibiting AKT pathway | [61] |
| Osteoarthritis | Regulates angiogenesis through inversely association of VEGF levels | [62] | ||
| Angiogenesis | Overexpressed MEG3 suppresses VEC proliferation, migration and angiogenesis via regulating miR-9 | [63] | ||
| Ischemic brain injury | Downregulated MEG3 promotes angiogenesis via negatively regulated Notch pathways | [64] | ||
| Cerebral infarction | Downregulated MEG3 promotes angiogenesis of OGD/R-induced rat brain microvascular endothelial cells (RBMVECs) through regulating P53/NOX4 axis | [65] | ||
| Angiogenesis | Downregulated MEG3 suppresses VEGF-induced EC migration and angiogenesis through modulating VEGFR2 expression | [66] | ||
| Linc00511 | 17q24.3 | Pancreatic ductal adenocarcinoma | Promotes tumour cells proliferation, migration, invasion and angiogenesis through sponging miR-29b-3p | [190] |
| Small nucleolar RNA host gene 15 (SNHG15) | 7p13 | Glioma | Promotes glioma VEC proliferation, migration and tube formation by increasing VEGFA and cell division cycle 42 (Cdc42) expression by targeting miR-153 | [191] |
| LincRNA-p21 | 6p21.2 | Non–Small Cell Lung Cancer | Enhances VEGFA production and promotes angiogenesis | [192] |
| ANRIL | 9p21.3 | Diabetes mellitus | Overexpressed ANRIL promotes angiogenesis by increasing VEGF expression via activating NF-kB pathway | [74] |
| HOTAIR | 12q13.13 | Nasopharyngeal carcinoma | Promotes angiogenesis through upregulating VEGFA and angiopoietin 2 (Ang2) expression by glucose regulated protein 78 (GRP78). | [76] |
| H19 | 11p15.5 | Glioma | Promotes glioma-induced angiogenesis by increasing miR-29a | [81] |
| LINC00657 | 20q11.23 | Angiogenesis | Promotes oxLDL-mediated EC proliferation, migration and tube formation by interacting with miR-590-3p and increasing VEGF, MMP-2 and MMP-9 expression | [193] |
| lncRNA | Correlations of the Functional Involvement of lncRNAs in ED and Diabetes | References |
|---|---|---|
| ANRIL | Up-regulated in DM and alters the EC function through increasing VEGF expression by P300/miR200b/EZH2 | [75] |
| MALAT1 | Highly expressed in DM and upregulates inflammatory mediators IL-6 & TNF-α through activating SAA3 and that stimulates DM-induced EC dysfunction via p38MAPK signaling pathway | [55,56,57] |
| MEG3 | Down-expressed in DM and enhances DM-mediated EC dysfunctions through altering PI3K/Akt signaling pathway | [67,68] |
| Myocardial infarction associated transcript (MIAT) | Induces DM induced EC dysfunction by acting as a competing endogenous RNA (CeRNA) via MIAT/miR-150-5p/VEGF network | [209] |
| RNCR3 | Up-regulated in DM and stimulates DM-induced retinal EC dysfunction through regulating RNCR3/ Kruppel like factor 2 (KLF2)/miR-185-5p pathway | [79] |
| lncRNA. | Correlations of the Functional Involvement of lncRNAs in ED and Atherosclerosis | References |
|---|---|---|
| Lnc00113 | Promotes abnormal EC proliferation, survival and migration via activating PI3K/Akt/mTOR pathway that disrupt EC function and develop atherosclerosis | [230] |
| MALAT1 | Protects EC from ox-LDL-induced EC dysfunction through inhibiting miR-22-3P and upregulating C-X-C Motif Chemokine Receptor 2 (CXCR2) & AKT expression in the settings of atherosclerosis | [58] |
| HOTAIR | Protect EC from injury and enhance EC proliferation, migration and inhibit apoptosis via thymic stromal lymphopoietin (TSLP)-PI3K/AKT-HOTAIR pathway that regulates angiogenesis pathogenesis and progression. | [77] |
| RP11-714G18.1 | Suppresses EC migration through RP11-714G18.1/ LRP2 binding protein (LRP2BP)/MMP1 signaling pathway that provide athero-defensive role in atherosclerosis-related EC dysfunction. | [231] |
| lincRNA-p21 | Stimulates ox-LDL-induced EC apoptosis and LOX-1 expression via activation of protein kinase C delta (PKCδ) that regulates atherosclerosis pathogenesis | [232] |
| TCONS_00024652 | Promotes EC proliferation, migration and angiogenesis via downregulating miR21 expression that stimulates atherosclerosis progression | [233] |
| XIST | Promotes ox-LDL-mediated EC injury via miR-320/ nucleotide binding oligomerization domain containing 2 (NOD2) pathway and modulates atherosclerosis | [80] |
| RNCR3 | Regulates EC function and accelerates atheroprotective function to the endothelium via acting as a ceRNA and forming a feedback loop with KLF2 and miR-185-5p | [78] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, R.; Lai, C. A Brief Overview of lncRNAs in Endothelial Dysfunction-Associated Diseases: From Discovery to Characterization. Epigenomes 2019, 3, 20. https://doi.org/10.3390/epigenomes3030020
Islam R, Lai C. A Brief Overview of lncRNAs in Endothelial Dysfunction-Associated Diseases: From Discovery to Characterization. Epigenomes. 2019; 3(3):20. https://doi.org/10.3390/epigenomes3030020
Chicago/Turabian StyleIslam, Rashidul, and Christopher Lai. 2019. "A Brief Overview of lncRNAs in Endothelial Dysfunction-Associated Diseases: From Discovery to Characterization" Epigenomes 3, no. 3: 20. https://doi.org/10.3390/epigenomes3030020