A Proposed Clinical Decision Support Architecture Capable of Supporting Whole Genome Sequence Information
Abstract
:1. Introduction
1.1. State of the Art
1.2. Technical Desiderata
| Desiderata Number | Desiderata Description |
|---|---|
| 1 | Maintain a separation of primary molecular observations from the clinical interpretations of those data |
| 2 | Support lossless data compression from primary molecular observations to clinically manageable subsets |
| 3 | Maintain the linkage of molecular observations to the laboratory methods used to generate them |
| 4 | Support a compact representation of clinically actionable subsets for optimal performance |
| 5 | Simultaneously support human-viewable formats and machine-readable formats in order to facilitate the implementation of decision support rules |
| 6 | Anticipate fundamental changes in the understanding of human molecular variation |
| 7 | Support both individual clinical care and discovery science |
| Desiderata Number | Desiderata Description |
|---|---|
| 8 | CDS knowledge must have the potential to incorporate multiple genes and clinical information |
| 9 | Keep CDS knowledge separate from variant classification |
| 10 | CDS knowledge must have the capacity to support multiple EHR platforms with various data representations with minimal modification |
| 11 | Support a large number of gene variants, while simplifying the CDS knowledge to the extent possible |
| 12 | Leverage current and developing CDS and genomics infrastructure and standards |
| 13 | Support a CDS knowledge base deployed at and developed by multiple independent organizations |
| 14 | Access and transmit only the genomic information necessary for CDS |
1.3. Study Objective
2. Methods
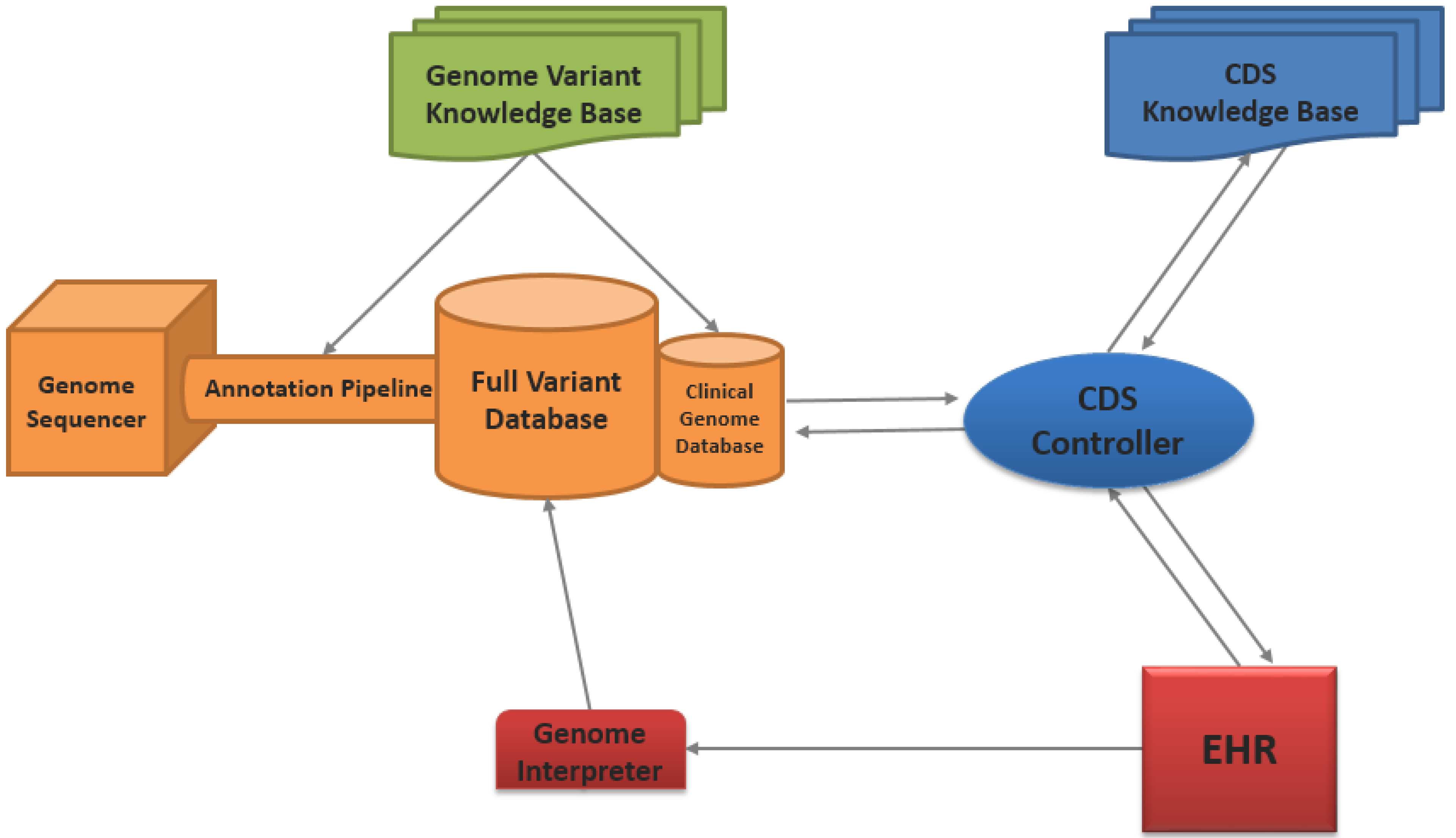
2.1. Architecture Overview
SOA CDS for WGS Information
2.2. Genome Sequencing and Annotation Pipeline
Genome Annotation
2.3. Genome Variant Knowledge Base
2.3.1. Variant Clinical Interpretations Categories
2.3.2. Variant Knowledge Management
2.4. Genome Databases
2.4.1. Genome Data Considerations
2.4.2. Database Approach
2.5. The Roles of the Electronic Health Record
2.5.1. EHR as a Repository of Clinical Data
2.5.2. CDS Interface with End Users
2.5.3. Leveraging Available EHR Capabilities
2.6. CDS Knowledge Base
CDS Knowledge Development and Management
2.7. CDS Controller
- The CDS controller obtains clinical data from the EHR in a standardized format (e.g., vMR), as a result of a CDS trigger within the EHR.
- The patient data is compared with the data requirements for the requested CDS knowledge module. In the case of our architecture, the CDS controller will identify that the patient’s genomic information required by CDS knowledge is missing and will make a request to the genome database for that information.
- The CDS controller obtains the patient’s genomic information from the clinical genome database, as specified by the CDS knowledge data requirements.
- The CDS controller then merges the patient’s genome information with the clinical information into a single vMR file.
- The complete data package is subsequently transmitted to the CDS knowledge base for evaluation.
- After CDS evaluation, the CDS controller receives the CDS response from the CDS knowledge base. At this point, the CDS controller can then process the CDS responses with additional workflow requirements (e.g., human review and approval of CDS recommendation), if necessary.
- The CDS response is relayed to the EHR for end-user presentation.
2.8. Genome Interpreter
3. Results
Meeting the Technical Desiderata
4. Discussion
4.1. Comparison of Proposed Architecture to Prior Work on CDS for Genomics
| WGS barriers | Desiderata requirements | How the proposed architecture addresses requirements |
|---|---|---|
| Clinical interpretations of genomic information can be dynamic [30] | (Desiderata #1) Maintain a separation of primary molecular observations from the clinical interpretations of those data | The genome variant knowledge bases exist separately and independently from the genome databases |
| WGS information contains a large amount of redundant and non-relevant data [38] | (Desiderata #2) Support lossless data compression from primary molecular observations to clinically manageable subsets | Genome variant file formats are based on a reference sequence, and a clinical genome database is used |
| Genomic results may be different based upon laboratory methods [72] | (Desiderata #3) Maintain a linkage of molecular observations to the laboratory methods used to generate them | Laboratory methods are included with the variant file in the full genome database |
| A majority of a patient’s 3,000,000+ genome variants will not have a clinical impact [4] | (Desiderata #4) Support the compact representation of clinically actionable subsets for optimal performance | Compact representation of clinically actionable informatics are available in the clinical genome database |
| Computing on the genome will require data representations that are hard for humans to understand [61] | (Desiderata #5) Simultaneously support human-viewable formats and machine-readable formats in order to facilitate the implementation of decision support rules | The machine-readable data format is used throughout the architecture, whereas a human viewable format is available through the genome interpreter |
| Our understanding of the human genome is nascent and may change significantly in the future [73] | (Desiderata #6) Anticipate fundamental changes in the understanding of human molecular variation | The proposed SOA architecture design allows for the flexibility of components to adapt to additional requirements as needed |
| Using available clinical and genomic information will be essential for research and discovery [74] | (Desiderata #7) Support both individual clinical care and discovery science | The same methods used to gather clinical and genomic data for CDS can be used for research, as well |
| Relatively few diseases are caused by a single genetic variant alone [75] | (Desiderata #8) CDS knowledge must have the potential to incorporate multiple genes and clinical information | The CDS controller is able to collect all required clinical and genomic data required by the CDS knowledge base |
| CDS knowledge may evolve independent of variant classifications [30] | (Desiderata #9) Keep CDS knowledge separate from variant classification | The CDS knowledge base is a separate component from the genome variant knowledge base |
| Many organizations, with various EHR platforms, will likely not be able to develop their own CDS for WGS information [65] | (Desiderata #10) CDS knowledge must have the capacity to support multiple EHR platforms with various data representations with minimal modification | The architecture uses industry standards and approaches for scalable, interoperable CDS that are being considered for inclusion in EHR certification criteria related to Meaningful Use Stage 3 |
| A single gene can have 100s–1,000s of variants with various clinical impacts [76] | (Desiderata #11) Support a large number of gene variants, while simplifying the CDS knowledge to the extent possible | The information in the clinical genome database and required for CDS can simply consist of the gene and its clinical interpretation |
| Re-inventing prior standards work on genomics and CDS just for this use case may prove to be futile [57] | (Desiderata #12) Leverage current and developing CDS and genomics infrastructure and standards | Health IT and genetics standards are used throughout the architecture |
| No single entity will be able to develop and maintain all possible CDS knowledge for WGS [69] | (Desiderata #13) Support a CDS knowledge base deployed at and developed by multiple independent organizations | Service-based CDS supports CDS knowledge developed and maintained by multiple, independent organizations |
| The file size and security concerns for WGS information are important [77] | (Desiderata #14) Access and transmit only the genomic information necessary for CDS | The CDS controller requests only the genome data needed for CDS knowledge |
Originality and Uniqueness of the Proposed Architecture
4.2. Barriers Still to Overcome
4.3. Current Efforts and Future Direction
5. Conclusions
Glossary of Key Terms Used
| CDS controller | A component of a SOA architecture which links several services together |
| CDS knowledge | A representation of clinical knowledge in the form of logic, rules, expressions, guidelines or algorithms |
| CDS knowledge base | A repository of CDS knowledge |
| Clinical genome database | A repository which stores only variants of known or potential clinical importance |
| Clinical interpretation | The clinical impact of a variant |
| Full genome database | A repository which stores all variants of an individual’s genome |
| Genome annotation | The process of locating and identifying key features of a genome |
| Genome interpreter | A visual interface which allows a clinician to manually review a patient’s genome variants |
| Genome sequencing | The process of obtaining the DNA sequence of an individual |
| Genome variant knowledge base | A repository of variants and associated clinical interpretation |
| Genome variant | A difference in a genome relative to a reference genome sequence |
| Service | A self-contained component with well-defined, understood capabilities |
| Service oriented architecture (SOA) | A software design methodology which contains several independent services |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- President’s Council of Advisors on Science and Technology. Priorities for Personalized Medicine; PCAST: Washington, DC, USA, 2008.
- Abrahams, E.; Ginsburg, G.S.; Silver, M. The personalized medicine coalition: Goals and strategies. Am. J. Pharmacogenomics 2005, 5, 345–355. [Google Scholar] [CrossRef]
- Bonetta, L. Whole-genome sequencing breaks the cost barrier. Cell 2010, 141, 917–919. [Google Scholar] [CrossRef]
- Ashley, E.A.; Butte, A.J.; Wheeler, M.T.; Chen, R.; Klein, T.E.; Dewey, F.E.; Dudley, J.T.; Ormond, K.E.; Pavlovic, A.; Morgan, A.A.; et al. Clinical assessment incorporating a personal genome. Lancet 2010, 375, 1525–1535. [Google Scholar] [CrossRef]
- Lupski, J.R.; Reid, J.G.; Gonzaga-Jauregui, C.; Rio Deiros, D.; Chen, D.C.Y.; Nazareth, L.; Bainbridge, M.; Dinh, H.; Jing, C.; Wheeler, D.A.; et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N. Engl. J. Med. 2010, 362, 1181–1191. [Google Scholar] [CrossRef]
- Rope, A.F.; Wang, K.; Evjenth, R.; Xing, J.; Johnston, J.J.; Swensen, J.J.; Johnson, W.E.; Moore, B.; Huff, C.D.; Bird, L.M.; et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am. J. Hum. Genet. 2011, 89, 28–43. [Google Scholar] [CrossRef]
- Talkowski, M.E.; Ordulu, Z.; Pillalamarri, V.; Benson, C.B.; Blumenthal, I.; Connolly, S.; Hanscom, C.; Hussain, N.; Pereira, S.; Picker, J.; et al. Clinical diagnosis by whole-genome sequencing of a prenatal sample. N. Engl. J. Med. 2012, 367, 2226–2232. [Google Scholar] [CrossRef]
- Wetterstrand, K. DNA sequencing costs: Data from the NHGRI genome sequencing program (GSP). Available online: http://www.genome.gov/sequencingcosts/ (accessed on 6 February 2013).
- Welch, B.M.; Kawamoto, K. The need for clinical decision support integrated with the electronic health record for the clinical application of whole genome sequencing information. J. Pers. Med. 2013, 3, 306–325. [Google Scholar] [CrossRef]
- Downing, G.J.; Boyle, S.N.; Brinner, K.M.; Osheroff, J.A. Information management to enable personalized medicine: Stakeholder roles in building clinical decision support. BMC Med. Inform. Decis. Mak. 2009, 9, e44. [Google Scholar] [CrossRef]
- Osheroff, J.A.; Teich, J.M.; Middleton, B.; Steen, E.B.; Wright, A.; Detmer, D.E. A roadmap for national action on clinical decision support. J. Am. Med. Inform. Assoc. 2007, 14, 141–145. [Google Scholar] [CrossRef]
- Wright, A.; Sittig, D.F.; Ash, J.S.; Feblowitz, J.; Meltzer, S.; McMullen, C.; Guappone, K.; Carpenter, J.; Richardson, J.; Simonaitis, L.; et al. Development and evaluation of a comprehensive clinical decision support taxonomy: Comparison of front-end tools in commercial and internally developed electronic health record systems. J. Am. Med. Inform. Assoc. 2011, 18, 232–242. [Google Scholar] [CrossRef]
- Bates, D.; Kuperman, G. Ten commandments for effective clinical decision support: Making the practice of evidence-based medicine a reality. J. Am. Med. Inform. Assoc. 2003, 10, 523–530. [Google Scholar] [CrossRef]
- Kawamoto, K.; Houlihan, C.A.; Balas, E.A.; Lobach, D.F. Improving clinical practice using clinical decision support systems: A systematic review of trials to identify features critical to success. Br. Med. J. 2005, 330, e765. [Google Scholar] [CrossRef]
- Welch, B.M.; Kawamoto, K. Clinical decision support for genetically guided personalized medicine: A systematic review. J. Am. Med. Inform. Assoc. 2012, 20, 388–400. [Google Scholar] [CrossRef]
- Drohan, B.; Ozanne, E.M.M.; Hughes, K.S.S. Electronic health records and the management of women at high risk of hereditary breast and ovarian cancer. Breast J. 2009, 15, S46–S55. [Google Scholar] [CrossRef]
- Tarczy-Hornoch, P.; Amendola, L.; Aronson, S.J.; Garraway, L.; Gray, S.; Grundmeier, R.W.; Hindorff, L.A.; Jarvik, G.; Karavite, D.; Lebo, M.; et al. A survey of informatics approaches to whole-exome and whole-genome clinical reporting in the electronic health record. Genet. Med. 2013, 15, 824–832. [Google Scholar] [CrossRef]
- Overby, C.L.; Kohane, I.; Kannry, J.L.; Williams, M.S.; Starren, J.; Bottinger, E.; Gottesman, O.; Denny, J.C.; Weng, C.; Tarczy-Hornoch, P.; et al. Opportunities for genomic clinical decision support interventions. Genet. Med. 2013, 15, 817–823. [Google Scholar] [CrossRef]
- Masys, D.R.; Jarvik, G.P.; Abernethy, N.F.; Anderson, N.R.; Papanicolaou, G.J.; Paltoo, D.N.; Hoffman, M.A.; Kohane, I.S.; Levy, H.P. Technical desiderata for the integration of genomic data into Electronic Health Records. J. Biomed. Inform. 2012, 45, 419–422. [Google Scholar] [CrossRef]
- Welch, B.M.; Eilbeck, K.; Del Fiol, G.; Meyer, L.; Kawamoto, K. Technical desiderata for the integration of genomic data with clinical decision support. 2014; submitted for publication. [Google Scholar]
- Starren, J.; Williams, M.S.; Bottinger, E.P. Crossing the omic chasm: A time for omic ancillary systems. JAMA 2013, 309, 1237–1238. [Google Scholar] [CrossRef]
- Erl, T. Service-Oriented Architecture (SOA): Concepts, Technology, and Design; Prentice Hall: Upper Saddle River, NJ, USA, 2005; pp. 1–792. [Google Scholar]
- He, H. What Is Service-Oriented Architecture? Available online: http://www.xml.com/pub/a/ws/2003/09/30/soa.html/ (accessed on 1 December 2013).
- Juneja, G.; Dournaee, B.; Natoli, J.; Birkel, S. Improving Performance of Healthcare Systems with Service Oriented Architecture. Available online: http://www.infoq.com/articles/soa-healthcare/ (accessed on 11 November 2013).
- Kawamoto, K.; Lobach, D.F. Design, implementation, use, and preliminary evaluation of SEBASTIAN, a standards-based Web service for clinical decision support. AMIA Annu. Symp. Proc. 2005, 2005, 380–384. [Google Scholar]
- Kawamoto, K.; Jacobs, J.; Welch, B.M.; Huser, V.; Paterno, M.D.; Del Fiol, G.; Shields, D.; Strasberg, H.R.; Haug, P.J.; Liu, Z.; et al. Clinical information system services and capabilities desired for scalable, standards-based, service-oriented decision support: Consensus assessment of the health level 7 clinical decision support work group. AMIA Annu. Symp. Proc. 2012, 2012, 446–455. [Google Scholar]
- Kawamoto, K.; Del Fiol, G.; Orton, C.; Lobach, D.F. System-agnostic clinical decision support services: Benefits and challenges for scalable decision support. Open Med. Inform. J. 2010, 4, 245–254. [Google Scholar] [CrossRef]
- Kawamoto, K.; Lobach, D. Proposal for fulfilling strategic objectives of the US roadmap for national action on decision support through a service-oriented architecture leveraging HL7 services. J. Am. Med. Inform. Assoc. 2007, 14, 146–155. [Google Scholar] [CrossRef]
- US Department of Health and Human Services. Voluntary 2015 Edition Electronic Health Record Certification Criteria: Interoperability Updates and Regulatory Improvements. Available online: http://www.regulations.gov/#!documentDetail;D=HHS-OS-2014-0002-0001/ (accessed on 12 March 2014).
- Aronson, S.J.; Clark, E.H.; Varugheese, M.; Baxter, S.; Babb, L.J.; Rehm, H.L. Communicating new knowledge on previously reported genetic variants. Genet. Med. 2012, 14, 713–719. [Google Scholar] [CrossRef]
- Kawamoto, K.; Lobach, D.F.; Willard, H.F.; Ginsburg, G.S. A national clinical decision support infrastructure to enable the widespread and consistent practice of genomic and personalized medicine. BMC Med. Inform. Decis. Mak. 2009, 9, e17. [Google Scholar] [CrossRef]
- Kawamoto, K. Integration of knowledge resources into applications to enable clinical decision support: Architectural considerations. In Clinical Decision Support: The Road Ahead; Greenes, R.A., Ed.; Elsevier: Burlington, MA, USA, 2007; pp. 503–538. [Google Scholar]
- Hamilton, A.B.; Oishi, S.; Yano, E.M.; Gammage, C.E.; Marshall, N.J.; Scheuner, M.T. Factors influencing organizational adoption and implementation of clinical genetic services. Genet. Med. 2014, 16, 238–245. [Google Scholar] [CrossRef]
- Wilcox, A.R.; Neri, P.M.; Volk, L.A.; Newmark, L.P.; Clark, E.H.; Babb, L.J.; Varugheese, M.; Aronson, S.J.; Rehm, H.L.; Bates, D.W. A novel clinician interface to improve clinician access to up-to-date genetic results. J. Am. Med. Inform. Assoc. 2014, 21, e117–e121. [Google Scholar] [CrossRef]
- Pulley, J.M.; Denny, J.C.; Peterson, J.F.; Bernard, G.R.; Vnencak-Jones, C.L.; Ramirez, A.H.; Delaney, J.T.; Bowton, E.; Brothers, K.; Johnson, K.; et al. Operational implementation of prospective genotyping for personalized medicine: The design of the vanderbilt PREDICT project. Clin. Pharmacol. Ther. 2012, 92, 87–95. [Google Scholar] [CrossRef]
- Gottesman, O.; Scott, S.; Ellis, S. The CLIPMERGE PGx program: Clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin. Pharmacol. Ther. 2013, 94, 214–217. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Reese, M.G.; Moore, B.; Batchelor, C.; Salas, F.; Cunningham, F.; Marth, G.T.; Stein, L.; Flicek, P.; Yandell, M.; Eilbeck, K. A standard variation file format for human genome sequences. Genome Biol. 2010, 11, R88. [Google Scholar] [CrossRef]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Howells, K.; Phillips, A.D.; Thomas, N.S.; Cooper, D.N. The Human Gene Mutation Database: 2008 update. Genome Med. 2009, 1, e13. [Google Scholar] [CrossRef]
- McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD). Online Mendelian Inheritance in Man, OMIM®. Available online: http://omim.org/ (accessed on 8 February 2013).
- National Center for Biotechnology Information. ClinVar. Available online: http://www.ncbi.nlm.nih.gov/clinvar/ (accessed on 8 February 2013).
- Fokkema, I.; Taschner, P.E.M.; Schaafsma, G.C.P.; Celli, J.; Laros, J.F.J.; den Dunnen, J.T. LOVD v.2.0: The next generation in gene variant databases. Hum. Mutat. 2011, 32, 557–563. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- Hu, H.; Huff, C.D.; Moore, B.; Flygare, S.; Reese, M.G.; Yandell, M. VAAST 2.0: Improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet. Epidemiol. 2013, 37, 622–634. [Google Scholar]
- Singleton, M.; Jorde, L.; Yandell, M. PHEVOR: Integration of VAAST with Phenomizer and the Gene Ontology for accurate disease-gene identification using only a single affected exome. In American Society of Human Genetics; University of Utah: Salt Lake City, UT, USA, 2013. [Google Scholar]
- Omicia. Omicia. Available online: http://www.omicia.com/ (accessed on 6 February 2013).
- SV Bio. SV Bio. Available online: http://svbio.com/ (accessed on 6 February 2013).
- Cypher Genomics. Available online: http://www.cyphergenomics.com/ (accessed on 1 December 2013).
- Wade, N. The quest for the $1,000 human genome: DNA sequencing in the doctor’s office? At birth? It may be coming closer. The New York Times, 18 July 2006; F1, F3. [Google Scholar]
- Cook-Deegan, R.; Conley, J.M.; Evans, J.P.; Vorhaus, D. The next controversy in genetic testing: Clinical data as trade secrets? Eur. J. Hum. Genet. 2013, 21, 585–588. [Google Scholar] [CrossRef]
- Riggs, E.R.; Wain, K.E.; Riethmaier, D.; Savage, M.; Smith-Packard, B.; Kaminsky, E.B.; Rehm, H.L.; Martin, C.L.; Ledbetter, D.H.; Faucett, W.A. Towards a universal clinical genomics database: The 2012 international standards for cytogenomic arrays consortium meeting. Hum. Mutat. 2013, 34, 915–919. [Google Scholar] [CrossRef]
- Richards, C.S.; Bale, S.; Bellissimo, D.B.; Das, S.; Grody, W.W.; Hegde, M.R.; Lyon, E.; Ward, B.E. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008, 10, 294–300. [Google Scholar] [CrossRef]
- Plon, S.; Eccles, D.; Easton, D. Sequence variant classification and reporting: Recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008, 29, 1282–1291. [Google Scholar] [CrossRef]
- Crews, K.R.; Gaedigk, A.; Dunnenberger, H.M.; Klein, T.E.; Shen, D.D.; Callaghan, J.T.; Kharasch, E.D.; Skaar, T.C. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin. Pharmacol. Ther. 2012, 91, 321–326. [Google Scholar] [CrossRef]
- The Human Cytochrome P450 (CYP) Allele Nomenclature Database. Available online: http://www.cypalleles.ki.se/ (accessed on 1 December 2013).
- Kho, A.N.; Rasmussen, L.V.; Connolly, J.J.; Peissig, P.L.; Starren, J.; Hakonarson, H.; Hayes, M.G. Practical challenges in integrating genomic data into the electronic health record. Genet. Med. 2013, 15, 772–778. [Google Scholar] [CrossRef]
- Gray, K.A.; Daugherty, L.C.; Gordon, S.M.; Seal, R.L.; Wright, M.W.; Bruford, E.A. Genenames.org: The HGNC resources in 2013. Nucleic Acids Res. 2013, 41, D545–D552. [Google Scholar] [CrossRef]
- Horaitis, O.; Cotton, R.G.H. The challenge of documenting mutation across the genome: The human genome variation society approach. Hum. Mutat. 2004, 23, 447–452. [Google Scholar] [CrossRef]
- Gymrek, M.; McGuire, A.L.; Golan, D.; Halperin, E.; Erlich, Y. Identifying personal genomes by surname inference. Science 2013, 339, 321–324. [Google Scholar] [CrossRef]
- Hoffman, M.A.; Williams, M.S. Electronic medical records and personalized medicine. Hum. Genet. 2011, 130, 33–39. [Google Scholar] [CrossRef]
- Huff, S.M. Ontologies, vocabularies, and data models. In Clinical Decision Support: The Road Ahead; Greenes, R.A., Ed.; Elsevier: Burlington, MA, USA, 2007; pp. 307–421. [Google Scholar]
- Kawamoto, K.; Del Fiol, G.; Strasberg, H.R.; Hulse, N.; Curtis, C.; Cimino, J.J.; Rocha, B.H.; Maviglia, S.; Fry, E.; Scherpbier, H.J.; et al. Multi-national, multi-institutional analysis of clinical decision support data needs to inform development of the HL7 virtual medical record standard. AMIA Annu. Symp. Proc. 2010, 2010, 377–381. [Google Scholar]
- Horsky, J.; Schiff, G.D.; Johnston, D.; Mercincavage, L.; Bell, D.; Middleton, B. Interface design principles for usable decision support: A targeted review of best practices for clinical prescribing interventions. J. Biomed. Inform. 2012, 45, 1202–1216. [Google Scholar] [CrossRef]
- Zhang, N.J.; Seblega, B.; Wan, T.; Unruh, L.; Agiro, A.; Miao, L. Health information technology adoption in US Acute care hospitals. J. Med. Syst. 2013, 37, e9907. [Google Scholar] [CrossRef]
- Zhang, M.; Velasco, F.; Musser, R.; Kawamoto, K. Enabling cross-platform clinical decision support through Web-based decision support in commercial electronic health record systems: Proposal and evaluation of initial prototype implementations. AMIA Annu. Symp. Proc. 2013, 2013, 1558–1567. [Google Scholar]
- Teich, J.M.; Osheroff, J.A.; Pifer, E.A.; Sittig, D.F.; Jenders, R.A. Clinical decision support in electronic prescribing: Recommendations and an action plan: Report of the joint clinical decision support workgroup. J. Am. Med. Inform. Assoc. 2005, 12, 365–376. [Google Scholar] [CrossRef]
- Kawamoto, K.; Del Fiol, G.; Lobach, D.F.; Jenders, R.A. Standards for scalable clinical decision support: Need, current and emerging standards, gaps, and proposal for progress. Open Med. Inform. J. 2010, 4, 235–244. [Google Scholar] [CrossRef]
- Gage, B.; Eby, C.; Johnson, J. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin. Pharmacol. Ther. 2008, 84, 326–331. [Google Scholar] [CrossRef]
- Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP working group: Genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet. Med. 2009, 11, 35–41. [Google Scholar] [CrossRef]
- Aronson, S.; Clark, E.; Babb, L. The geneinsight suite: A platform to support laboratory and provider use of DNA based genetic testing. Hum. Mutat. 2011, 32, 532–536. [Google Scholar] [CrossRef]
- Schrijver, I.; Aziz, N.; Farkas, D.H.; Furtado, M.; Gonzalez, A.F.; Greiner, T.C.; Grody, W.W.; Hambuch, T.; Kalman, L.; Kant, J.A.; et al. Opportunities and challenges associated with clinical diagnostic genome sequencing: A report of the association for molecular pathology. J. Mol. Diagn. 2012, 14, 525–540. [Google Scholar] [CrossRef]
- Ast, G. The alternative genome. Sci. Am. 2005, 292, 40–47. [Google Scholar]
- Gottesman, O.; Kuivaniemi, H.; Tromp, G.; Faucett, W.A.; Li, R.; Manolio, T.A.; Sanderson, S.C.; Kannry, J.; Zinberg, R.; Basford, M.A.; et al. The electronic medical records and genomics (eMERGE) network: Past, present, and future. Genet. Med. 2013, 15, 761–771. [Google Scholar] [CrossRef]
- Imamura, M.; Maeda, S. Genetics of type 2 diabetes: The GWAS era and future perspectives. Endocr. J. 2011, 58, 723–739. [Google Scholar] [CrossRef]
- Cystic Fibrosis Mutation Database: Statistics. Available online: http://www.genet.sickkids.on.ca/StatisticsPage.html/ (accessed on 19 February 2013).
- US Department of Health and Human Services, Office of the Secretary. Standards for Privacy of Individually Identifiable Health Information. In Final Rule; SEC: Washington, DC, USA, 2000; Volume 502, p. 45. [Google Scholar]
- Peterson, J.F.; Bowton, E.; Field, J.R.; Beller, M.; Mitchell, J.; Schildcrout, J.; Gregg, W.; Johnson, K.; Jirjis, J.N.; Roden, D.M.; et al. Electronic health record design and implementation for pharmacogenomics: A local perspective. Genet. Med. 2013, 15, 833–841. [Google Scholar] [CrossRef]
- Bell, G.C.; Crews, K.R.; Wilkinson, M.R.; Haidar, C.E.; Hicks, J.K.; Baker, D.K.; Kornegay, N.M.; Yang, W.; Cross, S.J.; Howard, S.C.; et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J. Am. Med. Inform. Assoc. 2014, 21, 93–99. [Google Scholar] [CrossRef]
- Hicks, J.K.; Crews, K.R.; Hoffman, J.M.; Kornegay, N.M.; Mark, R.; Lorier, R.; Stoddard, A.; Yang, W.; Smith, C.; Christian, A.; et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin. Pharmacol. Ther. 2012, 92, 563–566. [Google Scholar] [CrossRef]
- Clinvar Standard Terms for Clinical Significance. Available online: ftp://ftp.ncbi.nlm.nih.gov/pub/GTR/standard_terms/Clinical_significance.txt/ (accessed on 1 December 2013).
- Meaningful Use Criteria and How to Attain Meaningful Use of EHRs. Available online: http://www.healthit.gov/providers-professionals/how-attain-meaningful-use/ (accessed on 1 December 2013).
- Standards & Interoperability (S&I) Framework. Health eDecisions Homepage. Available online: http://wiki.siframework.org/Health+eDecisions+Homepage/ (accessed on 8 November 2013).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Welch, B.M.; Loya, S.R.; Eilbeck, K.; Kawamoto, K. A Proposed Clinical Decision Support Architecture Capable of Supporting Whole Genome Sequence Information. J. Pers. Med. 2014, 4, 176-199. https://doi.org/10.3390/jpm4020176
Welch BM, Loya SR, Eilbeck K, Kawamoto K. A Proposed Clinical Decision Support Architecture Capable of Supporting Whole Genome Sequence Information. Journal of Personalized Medicine. 2014; 4(2):176-199. https://doi.org/10.3390/jpm4020176
Chicago/Turabian StyleWelch, Brandon M., Salvador Rodriguez Loya, Karen Eilbeck, and Kensaku Kawamoto. 2014. "A Proposed Clinical Decision Support Architecture Capable of Supporting Whole Genome Sequence Information" Journal of Personalized Medicine 4, no. 2: 176-199. https://doi.org/10.3390/jpm4020176




