Implications of Possible HBV-Driven Regulation of Gene Expression in Stem Cell-like Subpopulation of Huh-7 Hepatocellular Carcinoma Cell Line
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Separation of Huh-7 CSCs
2.3. Plasmid Transfection
2.4. Invasion Assay
2.5. PCR Analyses for EMT Markers
2.6. miRNome Analysis
2.7. RNA-Seq Data Analysis
2.8. Phylogenetic Analysis of miRNAs
2.9. Genomic Database Analysis
2.10. Putative Protein Structure Homology Analysis
2.11. Cancer Cell Line Database Analysis
2.12. Statistics
3. Results
3.1. miRNA Profile upon Orai1 and STIM1 Overexpression
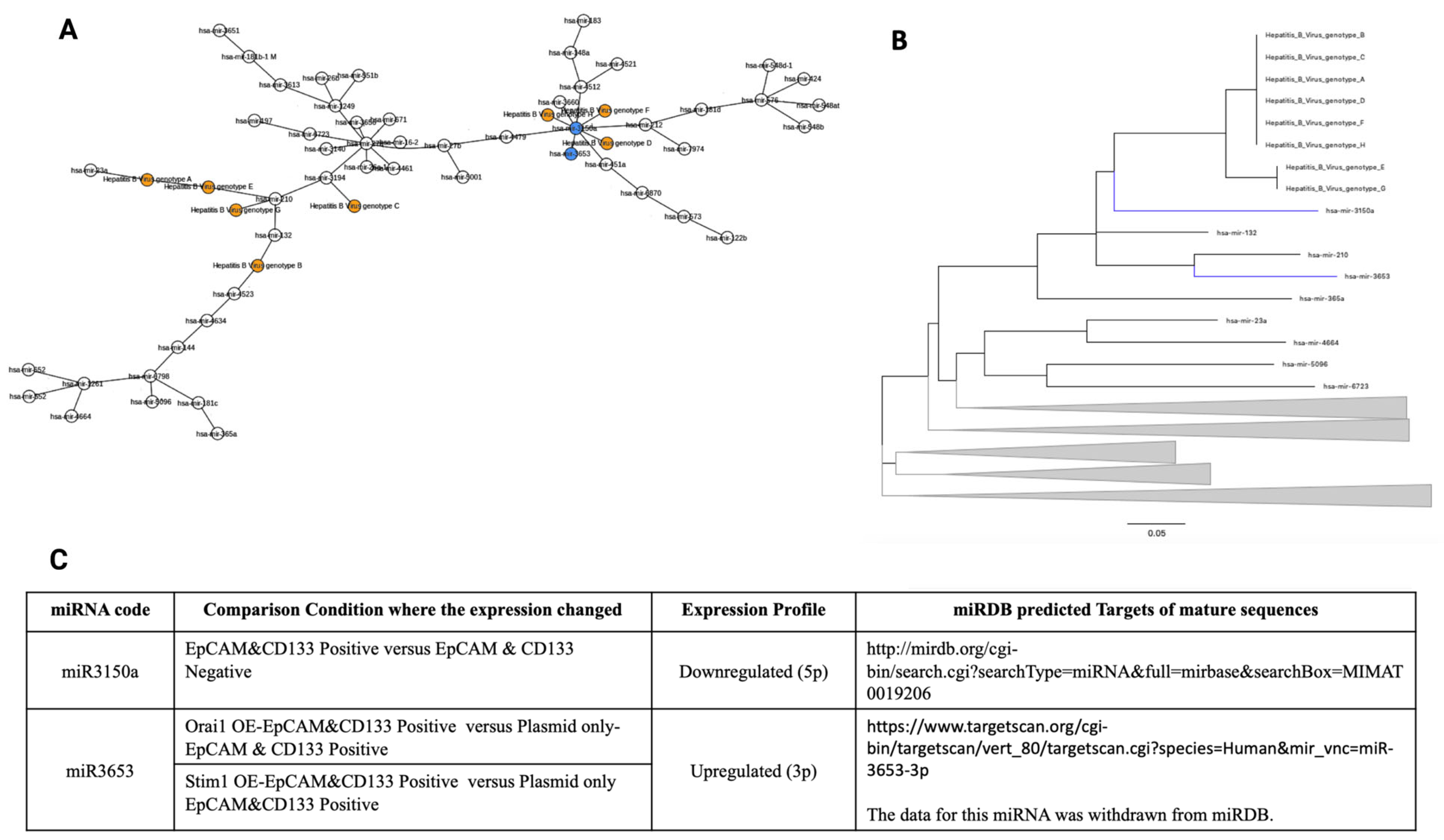
3.2. Phylogenetic Analysis of miRNAs
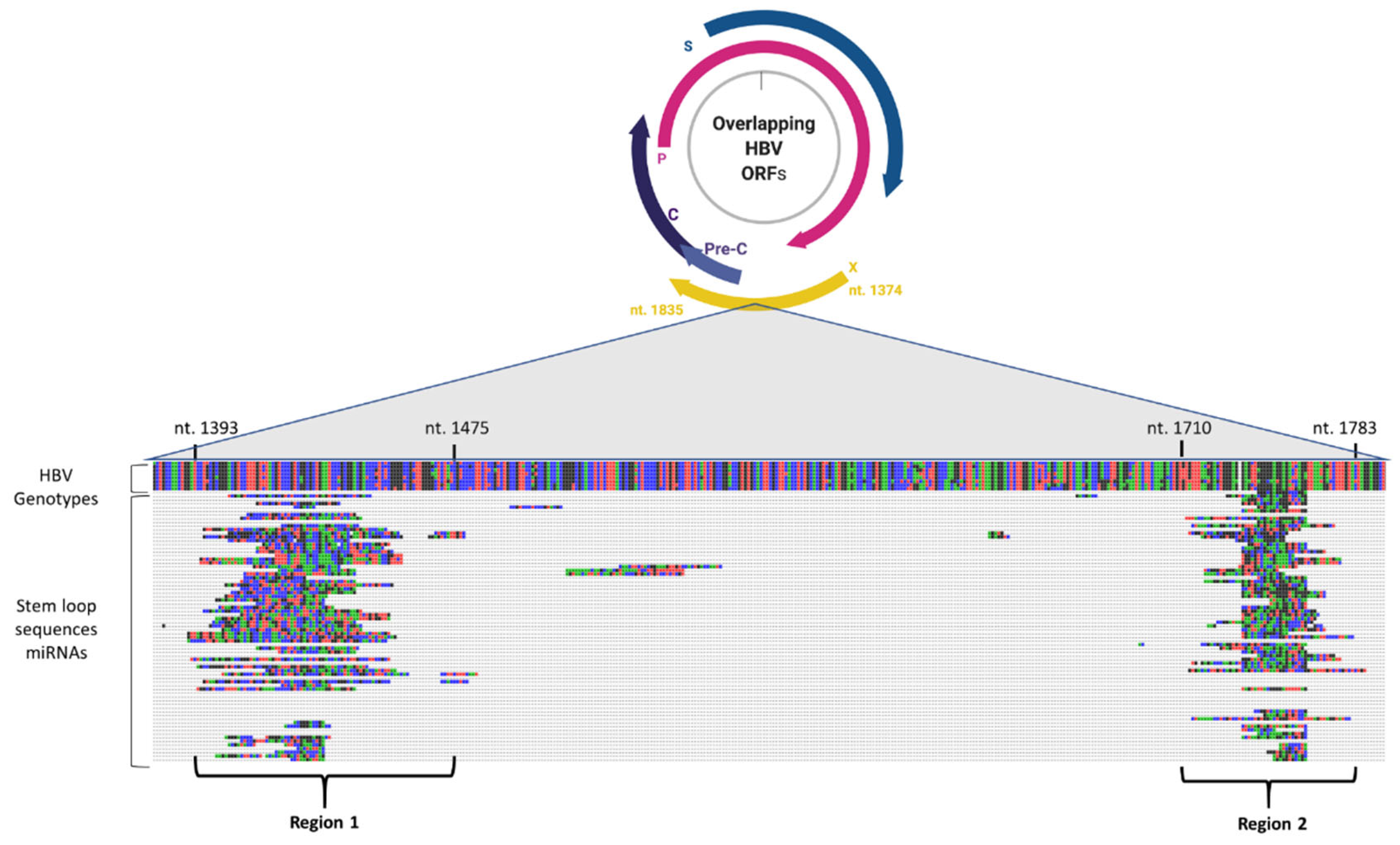
3.3. Genomic Database Analysis of Putative HBV Regions
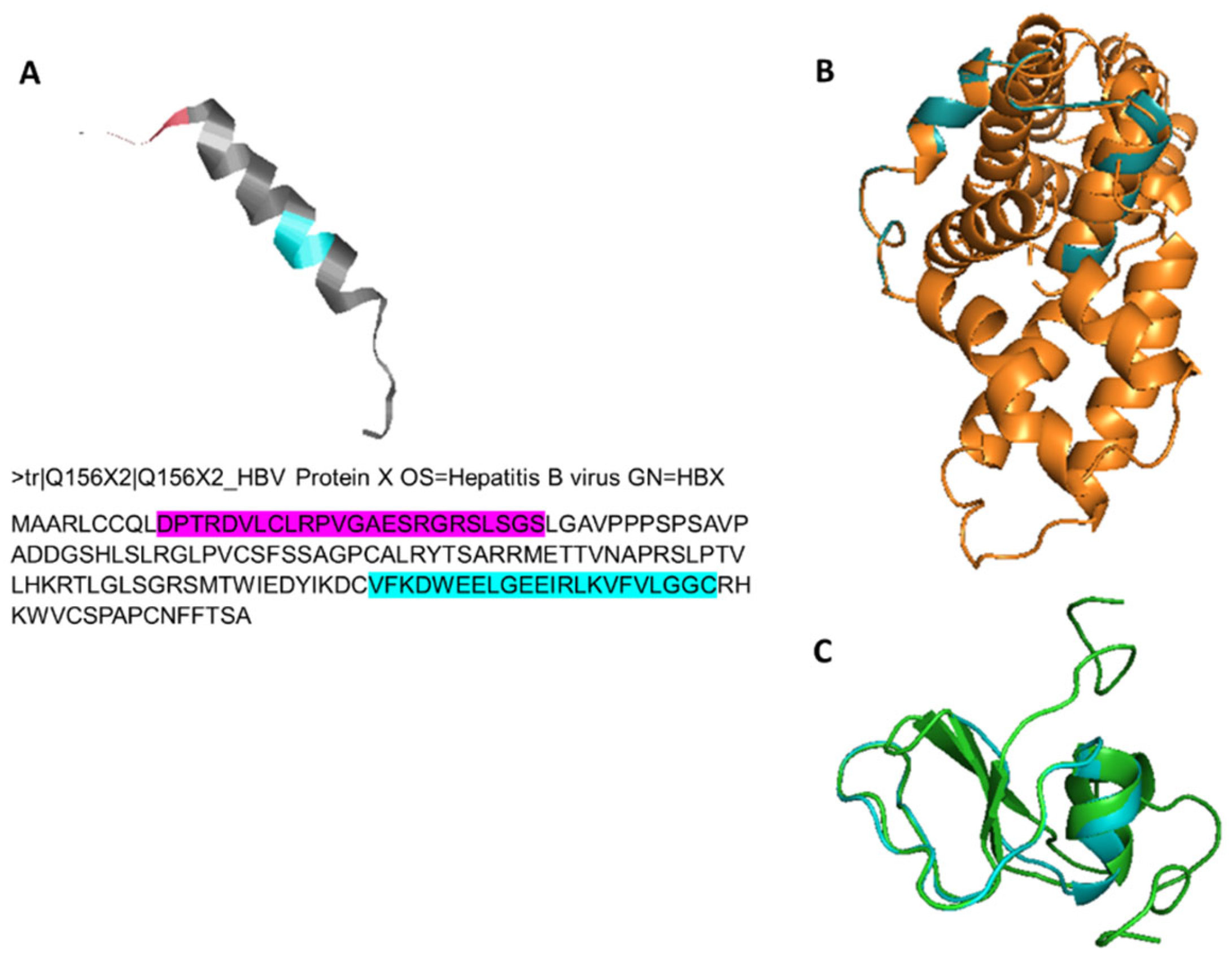
3.4. Structural Homology Analysis
3.5. E-Cadherin Expression Changes
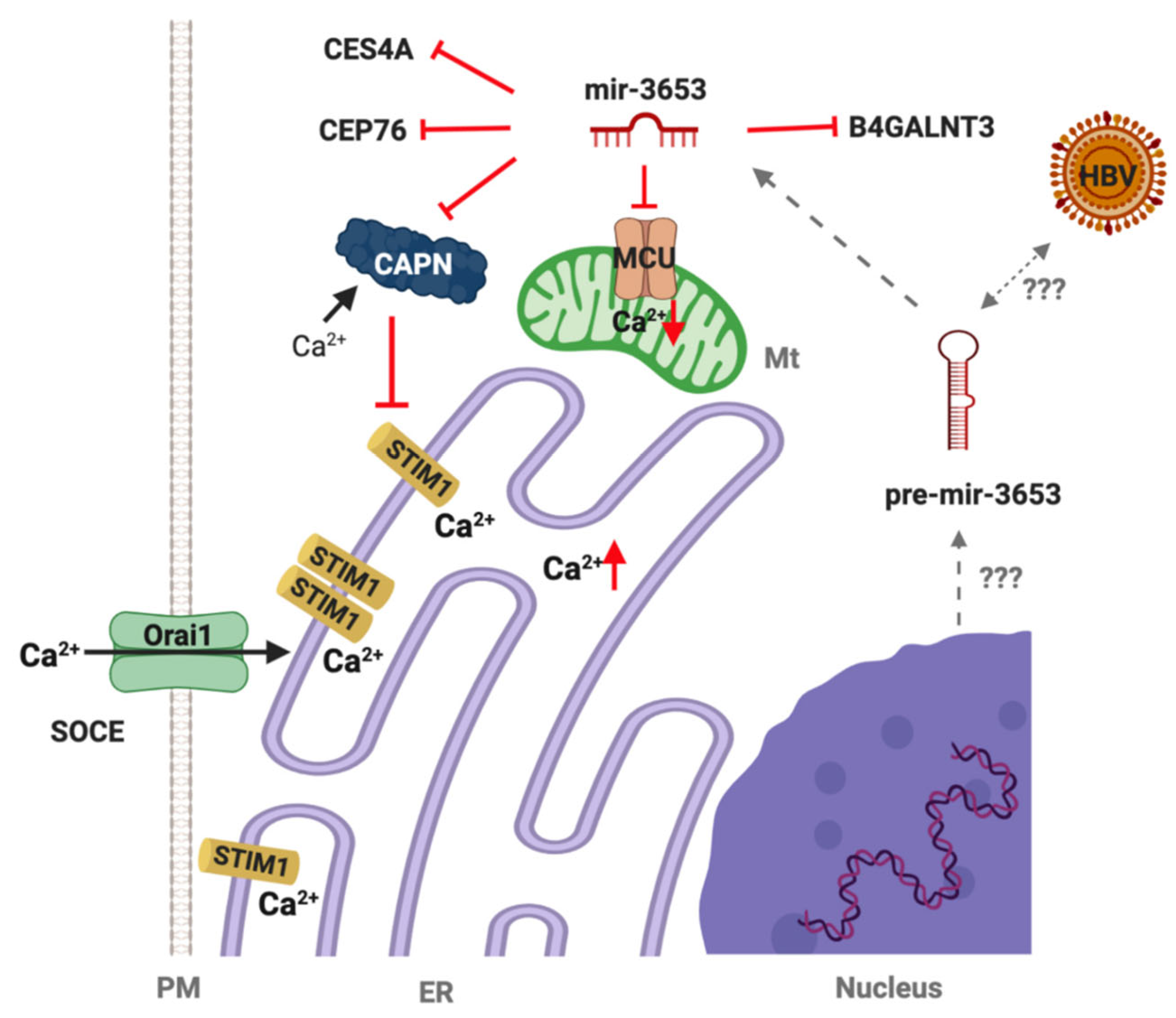
3.6. Expression Patterns of miR3653 Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liang, T.J. Hepatitis B: The virus and disease. Hepatology 2009, 49, S13–S21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, W.; Zhang, Z.; Ling, C.; Zheng, W.; Zhu, C.; Carr, M.J.; Higgins, D.G. Hepatitis B virus subgenotyping: History, effects of recombination, misclassifications, and corrections. Infect. Genet. Evol. 2013, 16, 355–361. [Google Scholar] [CrossRef]
- Tu, T.; Budzinska, M.A.; Shackel, N.A.; Urban, S. HBV DNA integration: Molecular mechanisms and clinical implications. Viruses 2017, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yu, D.; Zhang, H.; He, H.; Zhang, C.; Zhao, W.; Sheng, R.-G. CD133+EpCAM+ phenotype possesses more characteristics of tumor initiating cells in hepatocellular carcinoma Huh7 Cells. Int. J. Biol. Sci. 2012, 8, 992–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, T.K.W.; Cheung, V.C.H.; Ng, I.O.L. Liver tumor-initiating cells as a therapeutic target for hepatocellular carcinoma. Cancer Lett. 2013, 338, 101–109. [Google Scholar] [CrossRef]
- Liu, R.; Shen, Y.; Nan, K.; Mi, B.; Wu, T.; Guo, J.; Li, M.; Lv, Y.; Guo, H. Association between expression of cancer stem cell markers and poor differentiation of hepatocellular carcinoma: A meta-analysis (PRISMA). Medicine 2015, 94, e1306. [Google Scholar] [CrossRef]
- Karacicek, B.; Erac, Y.; Tosun, M. Functional consequences of enhanced expression of STIM1 and Orai1 in Huh-7 hepatocellular carcinoma tumor-initiating cells. BMC Cancer 2019, 19, 751. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Bouchard, M.J. The Hepatitis B Virus X Protein Elevates Cytosolic Calcium Signals by Modulating Mitochondrial Calcium Uptake. J. Virol. 2012, 86, 313–327. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Liu, Z.; Yi, J.; Wang, J.; Liu, Y. Hepatitis B Virus X Protein Upregulates Intracellular Calcium Signaling by Binding C-terminal of Orail Protein. Curr. Med. Sci. 2018, 38, 26–34. [Google Scholar] [CrossRef]
- Choi, Y.; Gyoo Park, S.; Yoo, J.H.; Jung, G. Calcium ions affect the hepatitis B virus core assembly. Virology 2005, 332, 454–463. [Google Scholar] [CrossRef]
- Paterlini-Bréchot, P.; Saigo, K.; Murakami, Y.; Chami, M.; Gozuacik, D.; Mugnier, C.; Lagorce, D.; Bréchot, C. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 2003, 22, 3911–3916. [Google Scholar] [CrossRef]
- Murata, M.; Matsuzaki, K.; Yoshida, K.; Sekimoto, G.; Tahashi, Y.; Mori, S.; Uemura, Y.; Sakaida, N.; Fujisawa, J.; Seki, T.; et al. Hepatitis B virus X protein shifts human hepatic transforming growth factor (TGF)-β signaling from tumor suppression to oncogenesis in early chronic hepatitis B. Hepatology 2009, 49, 1203–1217. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, Z.; Huang, P. The function of miRNAs in hepatocarcinogenesis induced by hepatitis B virus X protein (Review). Oncol. Rep. 2017, 38, 652–664. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Li, J.; Feng, G.; Wang, Y.; Yang, G.; Liu, Y.; Zhang, S.; Feng, J.; Zheng, X. Hepatitis B virus X protein enhances hepatocarcinogenesis by depressing the targeting of NUSAP1 mRNA by miR-18b. Cancer Biol. Med. 2019, 16, 276–287. [Google Scholar]
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and Host Interactions. Annu. Rev. Microbiol. 2010, 64, 123–141. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhao, J.J.; Wang, C.M.; Li, M.Y.; Han, P.; Wang, L.; Cheng, Y.-G.; Zoulim, F.; Ma, X.; Xu, D.-P. Altered expression profiles of microRNAs in a stable hepatitis B virus-expressing cell line. Chin. Med. J. 2009, 122, 10–14. [Google Scholar]
- Wang, J.; Shen, J.; Zhao, K.; Hu, J.; Dong, J.; Sun, J. STIM1 overexpression in hypoxia microenvironment contributes to pancreatic carcinoma progression. Cancer Biol. Med. 2019, 16, 100–108. [Google Scholar]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [Green Version]
- Caruso, S.; Calatayud, A.-L.; Pilet, J.; La Bella, T.; Rekik, S.; Imbeaud, S.; Letouzé, E.; Meunier, L.; Bayard, Q.; Rohr-Udilova, N.; et al. Analysis of Liver Cancer Cell Lines Identifies Agents With Likely Efficacy Against Hepatocellular Carcinoma and Markers of Response. Gastroenterology 2019, 157, 760–776. [Google Scholar] [CrossRef] [PubMed]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma′ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016, baw100. [Google Scholar] [CrossRef] [PubMed]
- Klijn, C.; Durinck, S.; Stawiski, E.W.; Haverty, P.M.; Jiang, Z.; Liu, H.; Degenhardt, J.; Mayba, O.; Gnad, F.; Liu, J.; et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2015, 33, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, T.; Deng, Z.; Sun, L. MicroRNA-3653 inhibits the growth and metastasis of hepatocellular carcinoma by inhibiting ITGB1. Oncol. Rep. 2019, 41, 1669–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podlaha, O.; Wu, G.; Downie, B.; Ramamurthy, R.; Gaggar, A.; Subramanian, M.; Ye, Z.; Jiang, Z. Genomic modeling of hepatitis B virus integration frequency in the human genome. PLoS ONE 2019, 14, e0220376. [Google Scholar] [CrossRef] [Green Version]
- Prins, D.; Michalak, M. STIM1 is cleaved by calpain. FEBS Lett. 2015, 589, 3294–3301. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Jiang, Z.; Wang, B.; Chang, L.; Liu, J.; Zhang, L.; Gu, L. Expression of STIM1 is associated with tumor aggressiveness and poor prognosis in breast cancer. Pathol. Res. Pract. 2017, 213, 1043–1047. [Google Scholar] [CrossRef]
- Udali, S.; Guarini, P.; Ruzzenente, A.; Ferrarini, A.; Guglielmi, A.; Lotto, V.; Tononi, P.; Pattini, P.; Moruzzi, S.; Campagnaro, T.; et al. DNA methylation and gene expression profiles show novel regulatory pathways in hepatocellular carcinoma. Clin. Epigenet. 2015, 7, 43. [Google Scholar] [CrossRef] [Green Version]
- Barbelanne, M.; Chiu, A.; Qian, J.; Tsang, W.Y. Opposing post-translational modifications regulate Cep76 function to suppress centriole amplification. Oncogene 2016, 35, 5377–5387. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Wang, L.; Wei, P.; Chi, Y.; Li, D.; Wang, Q.; Ni, S.; Tan, C.; Sheng, W.; Sun, M.; et al. Role of MUC20 overexpression as a predictor of recurrence and poor outcome in colorectal cancer. J. Transl. Med. 2013, 11, 151. [Google Scholar] [CrossRef] [Green Version]
- Kasprzak, A.; Adamek, A. Mucins: The old, the new and the promising factors in hepatobiliary carcinogenesis. Int. J. Mol. Sci. 2019, 20, 1288. [Google Scholar] [CrossRef] [Green Version]
- Katyal, S.; Oliver, J.H.; Peterson, M.S.; Ferris, J.V.; Carr, B.S.; Baron, L.R. Extrahepatic metastases of hepatocellular carcinoma. Radiology 2000, 216, 698–703. [Google Scholar] [CrossRef]
- Che, M.I.; Huang, J.; Hung, J.S.; Lin, Y.C.; Huang, M.J.; Lai, H.S.; Hsu, W.M.; Liang, J.-T.; Huang, M.C. β1, 4-N-acetylgalactosaminyltransferase III modulates cancer stemness through EGFR signaling pathway in colon cancer cells. Oncotarget 2014, 5, 3673–3684. [Google Scholar] [CrossRef]
- Huo, T.I.; Wang, X.W.; Forgues, M.; Wu, C.G.; Spillare, E.A.; Giannin, C.; Brechot, C.; Harris, C.C. Hepatitis B virus X mutants derived from human hepatocellular carcinoma retain the ability to abrogate p53-induced apoptosis. Oncogene 2001, 20, 3620–3628. [Google Scholar] [CrossRef] [Green Version]
- Chau, D.K.F.; Chen, G.G.; Zhang, H.; Leung, B.C.S.; Chun, S.; Lai, P.B.-S. Differential Functions of C- and N-Terminal Hepatitis B x Protein in Liver Cells Treated with Doxorubicin in Normoxic or Hypoxic Condition. PLoS ONE 2012, 7, e50118. [Google Scholar] [CrossRef] [Green Version]
- Quarleri, J. Core promoter: A critical region where the hepatitis B virus makes decisions. World J. Gastroenterol. 2015, 20, 425–435. [Google Scholar] [CrossRef]
- Meier-Stephenson, V.; Bremner, W.T.R.; Dalton, C.S.; van Marle, G.; Coffin, C.S.; Patel, T.R. Comprehensive Analysis of Hepatitis B Virus Promoter Region Mutations. Viruses 2018, 10, 603. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Mertz, J.E. Promoters for synthesis of the pre-C and pregenomic mRNAs of human hepatitis B virus are genetically distinct and differentially regulated. J. Virol. 1996, 70, 8719–8726. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.H.; Liu, X.; Yan, H.X.; Li, W.Y.; Zeng, X.; Yang, Y.; Zhao, J.; Liu, S.P.; Zhuang, X.-H.; Lin, C.; et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat. Commun. 2016, 7, 12992. [Google Scholar] [CrossRef] [Green Version]
- Furuta, M.; Tanaka, H.; Shiraishi, Y.; Unida, T.; Imamura, M.; Fujimoto, A.; Fujita, M.; Sasaki-Oku, A.; Maejima, K.; Nakano, K.; et al. Characterization of HBV integration patterns and timing in liver cancer and HBV-infected livers. Oncotarget 2018, 9, 25075–25088. [Google Scholar] [CrossRef] [Green Version]
- Budzinska, M.A.; Shackel, N.A.; Urban, S.; Tu, T. Cellular genomic sites of hepatitis B virus DNA integration. Genes 2018, 9, 365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, T.A.; Jiang, W.G. Loss of tight junction barrier function and its role in cancer metastasis. Biochim. Biophys. Acta Biomembr. 2009, 1788, 872–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, T.A.; Watkins, G.; Mansel, R.E.; Jiang, W.G. Loss of tight junction plaque molecules in breast cancer tissues is associated with a poor prognosis in patients with breast cancer. Eur. J. Cancer 2004, 40, 2717–2725. [Google Scholar] [CrossRef]
- Stucke, V.M.; Timmerman, E.; Vandekerckhove, J.; Gevaert, K.; Hall, A. The MAGUK protein MPP7 binds to the polarity protein hDlg1 and facilitates epithelial tight junction formation. Mol. Biol. Cell. 2007, 18, 1744–1755. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Bueno, G.; Portillo, F.; Cano, A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene 2008, 27, 6958–6969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spaderna, S.; Schmalhofer, O.; Wahlbuhl, M.; Dimmler, A.; Bauer, K.; Sultan, A.; Hlubek, F.; Jung, A.; Strand, D.; Eger, A.; et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008, 68, 123–141. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.Y.; Fattet, L.; Tsai, J.H.; Kajimoto, T.; Chang, Q.; Newton, A.C.; Yang, J. Apical–basal polarity inhibits epithelial–mesenchymal transition and tumour metastasis by PAR-complex-mediated SNAI1 degradation. Nat. Cell Biol. 2019, 21, 359–371. [Google Scholar] [CrossRef]
- Treyer, A.; Müsch, A. Hepatocyte polarity. Compr. Physiol. 2013, 3, 243–287. [Google Scholar]
- Fransvea, E.; Angelotti, U.; Antonaci, S.; Giannelli, G. Blocking transforming growth factor-beta up-regulates E-cadherin and reduces migration and invasion of hepatocellular carcinoma cells. Hepatology 2008, 47, 1557–1566. [Google Scholar] [CrossRef]
- Fransvea, E.; Mazzocca, A.; Antonaci, S.; Giannelli, G. Targeting transforming growth factor (TGF)-βRI inhibits activation of β1 integrin and blocks vascular invasion in hepatocellular carcinoma. Hepatology 2009, 49, 839–850. [Google Scholar] [CrossRef]
- Zhang, S.; Miao, Y.; Zheng, X.; Gong, Y.; Zhang, J.; Zhou, F.; Cai, C. STIM1 and STIM2 differently regulate endogenous Ca2+ entry and promote TGF-β-induced EMT in breast cancer cells. Biochem. Biophys. Res. Commun. 2017, 488, 74–80. [Google Scholar] [CrossRef]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The Three Es of Cancer Immunoediting. Annu. Rev. Immunol. 2004, 22, 329–360. [Google Scholar] [CrossRef]
- Sun, D.; Nassal, M. Stable HepG2- and Huh7-based human hepatoma cell lines for efficient regulated expression of infectious hepatitis B virus. J. Hepatol. 2006, 45, 636–645. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demir, A.B.; Benvenuto, D.; Karacicek, B.; Erac, Y.; Spoto, S.; Angeletti, S.; Ciccozzi, M.; Tosun, M. Implications of Possible HBV-Driven Regulation of Gene Expression in Stem Cell-like Subpopulation of Huh-7 Hepatocellular Carcinoma Cell Line. J. Pers. Med. 2022, 12, 2065. https://doi.org/10.3390/jpm12122065
Demir AB, Benvenuto D, Karacicek B, Erac Y, Spoto S, Angeletti S, Ciccozzi M, Tosun M. Implications of Possible HBV-Driven Regulation of Gene Expression in Stem Cell-like Subpopulation of Huh-7 Hepatocellular Carcinoma Cell Line. Journal of Personalized Medicine. 2022; 12(12):2065. https://doi.org/10.3390/jpm12122065
Chicago/Turabian StyleDemir, Ayse Banu, Domenico Benvenuto, Bilge Karacicek, Yasemin Erac, Silvia Spoto, Silvia Angeletti, Massimo Ciccozzi, and Metiner Tosun. 2022. "Implications of Possible HBV-Driven Regulation of Gene Expression in Stem Cell-like Subpopulation of Huh-7 Hepatocellular Carcinoma Cell Line" Journal of Personalized Medicine 12, no. 12: 2065. https://doi.org/10.3390/jpm12122065




