The Informational Substrate of Chemical Evolution: Implications for Abiogenesis
Abstract
1. Introduction: Stating the Problem
1.1. Chemical Evolution
1.2. Evolution of Autonomous Chemical Systems (ACSs)
1.3. Is There a Relationship between Chemical Evolution and Information Processing?
2. Chemical Information, Meaning and Interpretation
2.1. Information and Meaning at the Molecular Level
2.2. The Bases of Physicochemical Semiosis
3. Chemosemiosis: Chemical Evolution Arising from an Informational Background
3.1. The Interest of Chemosemiotic Models
3.2. Molecular Mechanisms of Information Processing in ACSs
4. Discussion
Funding
Acknowledgments
Conflicts of Interest
References
- Ponnamperuma, C. Chemical evolution and the origin of life. Nature 1964, 201, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Mirazo, K.; Briones, C.; de la Escosura, A. Prebiotic systems chemistry: New perspectives for the origins of life. Chem. Rev. 2014, 114, 285–366. [Google Scholar] [CrossRef] [PubMed]
- Ashkenasy, G.; Hermans, T.M.; Otto, S.; Taylor, A.F. Systems chemistry. Chem. Soc. Rev. 2017, 46, 2543–2554. [Google Scholar] [CrossRef] [PubMed]
- Vasas, V.; Szathmary, E.; Santos, M. Lack of evolvability in self-sustaining autocatalytic networks constraints metabolism-first scenarios for the origin of life. Proc. Natl. Acad. Sci. USA 2010, 107, 1470–1475. [Google Scholar] [CrossRef] [PubMed]
- Vasas, V.; Fernando, C.; Santos, M.; Kauffman, S.; Szathmary, E. Evolution before genes. Biol. Direct 2012, 7, 1. [Google Scholar] [CrossRef]
- Chen, I.A.; Nowak, M.A. From prelife to life: How chemical kinetcis become evolutionary dynamics. Acc. Chem. Res. 2012, 45, 2088–2096. [Google Scholar] [CrossRef] [PubMed]
- Szathmary, E.; Santos, M.; Fernando, C. Evolutionary potential and requirements for minimal protocells. Top. Curr. Chem. 2005, 259, 167–211. [Google Scholar]
- Pross, A. What is Life? How Chemistry Becomes Biology; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Pascal, R.; Pross, A.; Sutherland, J.D. Towards an evolutionary theory of the origin of life based on kinetics and thermodynamics. Open Biol. 2013, 3, 130156. [Google Scholar] [CrossRef]
- Van Esch, J.H.; Kaljn, R.; Otto, S. Chemical Systems Out of Equilibrium. Chem. Soc. Rev. 2017, 46, 5474–5475. [Google Scholar] [CrossRef]
- Mariani, A.; Sutherland, J.D. Non-enzymatic RNA backbone proofreading through energy-dissipative recycling. Angew. Chem. Int. Ed. 2017, 56, 6563–6566. [Google Scholar] [CrossRef]
- Piedrafita, G.; Monnard, P.A.; Mavelli, F.; Ruiz-Mirazo, K. Permeability-driven selection in a semi-empirical protocell model: The roots of prebiotic systems evolution. Sci. Rep. 2017, 7, 3141. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, K.; Tamura, M.; Shohda, K.I.; Toyota, T.; Suzuki, K.; Sugawara, T. Self-reproduction of supramolecular giant vesicles combined with the amplification of encapsulated DNA. Nat. Chem. 2011, 3, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Adamala, K.P.; Szostak, J.W. Nonenzymatic template-directed RNA synthesis inside model protocells. Science 2013, 342, 1098–1100. [Google Scholar] [CrossRef] [PubMed]
- Engelhart, A.E.; Adamala, K.P.; Szostak, J.W. A simple physical mechanism enables homeostasis in primitive cells. Nat. Chem. 2016, 8, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Ganti, T. Organization of chemical reactions into dividing and metabolizing units: The chemotons. Biosystems 1975, 7, 15–21. [Google Scholar] [CrossRef]
- Ganti, T. The Principles of Life; Oxford University Press: Oxford, UK, 2003. [Google Scholar]
- De la Escosura, A.; Briones, C.; Ruiz-Mirazo, K. The systems perspective at the crossroads between chemistry and biology. J. Biol. 2015, 381, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Mirazo, K.; Briones, C.; de la Escosura, A. Chemical roots of biological evolution: The origins of life as a process of development of autonomous functional systems. Open. Biol. 2017, 7, 170050. [Google Scholar] [CrossRef]
- Mossio, M.; Saborido, C.; Moreno, A. An organizational account of biological functions. Br. J. Philos. Sci. 2009, 60, 813–841. [Google Scholar] [CrossRef]
- Maturana, H.; Varela, F.J. Autopoiesis and Cognition. The Realization of the Living; Reidel Publishing Company: Dordrecht, The Netherlands, 1980. [Google Scholar]
- Ruiz-Mirazo, K.; Moreno, A. Basic autonomy as a fundamental step in the synthesis of life. Artif. Life 2004, 10, 235–259. [Google Scholar] [CrossRef]
- Boel, G.; Danot, O.; de Lorenzo, V.; Danchin, A. Omnipresent Maxwell´s demons orchestrate information management in living cells. Microb. Biotechnol. 2019, 12, 210–242. [Google Scholar] [CrossRef]
- Lehn, J.M. Supramolecular chemistry: Where from? Where to? Chem. Soc. Rev. 2017, 46, 2378–2379. [Google Scholar] [CrossRef] [PubMed]
- Mehta, E.L.; Lang, A.H.; Schqab, D.J. Landauer in the age of synthetic biology: Energy consumption and information processing in biochemical networks. J. Stat. Phys. 2016, 162, 1153–1166. [Google Scholar] [CrossRef]
- Hoffmann, P.M. Life´s Ratchet: How Molecular Machines Extract Order from Chaos; Basic Books: New York, NY, USA, 2012. [Google Scholar]
- Maxwell, J.C. Theory of Heat; Longmans, Green & Co.: London, UK, 1888. [Google Scholar]
- Hateren, J.H. What does Maxwell´s demon want from life? When information becomes functional and physical. arXiv 2015, arXiv:1407.8314. [Google Scholar]
- Deacon, T.W. Shanon-Boltzman-Darwin: Redefining information. Part 1. Cogn. Semiot. 2007, 1, 123–148. [Google Scholar]
- Deacon, T.W. Shanon-Boltzman-Darwin: Redefining information. Part 2. Cogn. Semiot. 2008, 2, 167–194. [Google Scholar]
- Maynard-Smith, J. The concept of information in biology. Philos. Sci. 2000, 67, 177–194. [Google Scholar] [CrossRef]
- Wills, P.R. DNA as information. Philos. Trans. A. 2016, 374, 2063. [Google Scholar] [CrossRef]
- Hoffmeyer, J. Biosemiotics. An Examination into the Science of Life and the Life of Science; University of Scranton Press: Scranton, PA, USA, 2008. [Google Scholar]
- Shannon, C. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 623–656. [Google Scholar] [CrossRef]
- Shannon, C.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Urbana, IL, USA, 1949. [Google Scholar]
- Seifert, U. Stochastic thermodynamics, fluctuation theorems, and molecular machines. Rep. Prog. Phys. 2012, 75, 126001. [Google Scholar] [CrossRef]
- Kuppers, B.O. Elements of a semantic code. In Evolution of Semantic Systems; Kuppers, B.O., Artmann, S., Hahn, U., Eds.; Springer-Verlag: Berlin/Heidelberg, UK, 2013; pp. 67–85. [Google Scholar]
- Kuppers, B.O. Information and the Origin of Life; The MIT Press: Cambridge, UK, 1990. [Google Scholar]
- Kuppers, B.O. The nucleation of semantic information in prebiotic matter. In Quasispecies: From Theory to Experimental Systems; Domingo, E., Schuster, P., Eds.; Springer International: Basel, Switzerland, 2016; pp. 23–42. [Google Scholar]
- Sharov, A.A. Functional information: Towards synthesis of biosemiotics and cybernetics. Entropy 2010, 12, 1050–1070. [Google Scholar] [CrossRef]
- Hendry, R.F. Is there downward causation in chemistry? In Philosophy of Chemistry. Synthesis of a New Discipline; Baird, D., Scerri, E., McIntyre, L., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 173–190. [Google Scholar]
- Emmeche, C. The computational notion of life. Theor. Segunda Epoca 1994, 9, 1–30. [Google Scholar]
- Emmeche, C.; Hoffmeyer, J. Code-duality and the semiotics of Nature. In On Semiotic Modelling; Anderson, M., Merrell, F., Eds.; De Gruyter Mouton: Berlin, Germany, 1991; pp. 117–166. [Google Scholar]
- Bay, Y.S.; Chotera, A.; Taran, O.; Liang, C.; Ashkenasy, G.; Lynn, D.G. Achieving biopolymer synergy in systems chemistry. Chem. Soc. Rev. 2018, 47, 5444–5456. [Google Scholar] [CrossRef]
- Cleaves, H.J.; Scott, A.M.; Hill, F.C.; Leszczynski, J.; Sahai, N.; Hazen, R. Mineral-organic interfacial processes: Potential roles in the origins of life. Chem. Soc. Rev. 2012, 41, 5502–5525. [Google Scholar] [CrossRef]
- Pizzarello, S.; Weber, A.L. Prebiotic amino acids as asymmetric catalysts. Science 2004, 303, 1151. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, R.; Adamala, K.; Gasperi, T.; Polticelli, F.; Stano, P. Small and random peptides: An unexplored reservoir of potentially functional primitive organocatalysts. The case of seryl-histidine. Life 2017, 7, 19. [Google Scholar] [CrossRef]
- Joyce, G.F.; Szostak, J.W. Protocells and RNA self-replication. Cold Spring Harb. Perspect. Med. 2018, 8, a034801. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.H.; Percivalle, C.; Ritson, D.J.; Duffy, C.D.; Sutherland, J.D. Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat. Chem. 2015, 7, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Bonfio, C.; Godino, E.; Corsini, M.; Fabrizi de Biani, F.; Guella, G.; Mansy, S.S. Prebiotic iron-sulfur peptide catalysts generate a pH gradient across model membranes of late protocells. Nat. Catal. 2018, 1, 616–623. [Google Scholar] [CrossRef]
- Krishnamurthy, R. Role of pKa of nucleobases in the origins of chemical evolution. Acc. Chem. Res. 2012, 45, 2035–2044. [Google Scholar] [CrossRef] [PubMed]
- Hordijk, W. Autocatalytic confusion clarified. J. Biol. 2017, 435, 22–28. [Google Scholar] [CrossRef]
- Paul, N.; Joyce, G.F. Minimal self-replicating systems. Curr. Opin. Chem. Biol. 2004, 8, 634–639. [Google Scholar] [CrossRef]
- Orgel, L.E. Self-organizing biochemical cycles. Proc. Natl. Acad. Sci. USA 2000, 97, 12503–12507. [Google Scholar] [CrossRef]
- Morowitz, H.J.; Kostelnik, J.D.; Yang, J.; Cody, G.D. The origin of intermediary metabolism. Proc. Natl. Acad. Sci. USA 2000, 97, 7704–7708. [Google Scholar] [CrossRef] [PubMed]
- Hordijk, W.; Kauffman, S.; Steel, M. Required levels of catalysis for emergence of autocatalytic sets in models of chemical reaction systems. Int. J. Mol. Sci. 2011, 12, 3085–3101. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, S.A. Autocatalytic sets of proteins. J. Biol. 1986, 119, 1–24. [Google Scholar]
- Decraene, J.; McMullin, B. The evolution of complexity in self-maintaining cellular information processing networks. Adv. Complex. Syst. 2011, 14, 55–75. [Google Scholar] [CrossRef]
- Adamala, K.; Szostak, J.W. Competition between model protocells driven by an encapsulated catalyst. Nat. Chem. 2013, 5, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Murillo-Sanchez, S.; Beaufils, D.; Mañas, J.M.G.; Pascal, R.; Ruiz-Mirazo, K. Fatty acids´ double role in the prebiotic formation of a hydrophobic dipeptide. Chem. Sci. 2016, 7, 3406–3413. [Google Scholar] [CrossRef] [PubMed]
- Drobot, B.; Iglesias-Artola, J.M.; Le Vay, K.; Kar, M.; Kreysing, M.; Mutschler, H.; Tang, T.Y.D. Compartmentalised RNA catalysis in membrane-free coacervate protocells. Nat. Commun. 2018, 9, 3643. [Google Scholar] [CrossRef]
- Stano, P.; Luisi, P.L. Semi-synthetic minimal cells: Origin and recent developments. Curr. Opin. Biotechnol. 2013, 24, 633–638. [Google Scholar] [CrossRef]
- Bissette, A.J.; Odell, B.; Fletcher, S.P. Physical autocatalysis driven by a bond-forming thiol-ene reaction. Nat. Commun. 2014, 5, 4607. [Google Scholar] [CrossRef]
- Lane, N.; Martin, W.F. The origin of membrane bioenergetics. Cell 2012, 151, 1406–1416. [Google Scholar] [CrossRef]
- Chen, I.A.; Szostak, J.W. Membrane growth can generate and transmembrane pH gradient in fatty acid vesicles. Proc. Natl. Acad. Sci. USA 2004, 101, 7965–7970. [Google Scholar] [CrossRef] [PubMed]
- Maiti, S.; Fortunati, I.; Ferrante, C.; Scrimin, P.; Prins, L.J. Dissipative self-assembly of vesicular nanoreactors. Nat. Chem. 2016, 8, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Kolchinsky, A.; Wolpert, D.H. Semantic information, autonomous agency and non-equilibrium statistical physics. Interface Focus 2018, 8, 20180041. [Google Scholar] [CrossRef] [PubMed]
- Hoffmeyer, J. Semiotic freedom. An emerging force. In Information and the Nature of Reality. From Physics to Metaphysics; Davies, P., Gregersen, N.H., Eds.; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- De Duve, C. Life Evolving: Molecules, Mind, and Meaning; Oxford University Press: Oxford, UK, 2002. [Google Scholar]
- Clayton, P.; Kauffman, S. On emergence, agency, and organization. Biol. Philos. 2006, 21, 501–521. [Google Scholar]
- Kauffman, S. The Origins of Order: Self-Organization and Selection in Evolution; Oxford University Press: Oxford, UK, 1993. [Google Scholar]


| Basic Function | Structure of Functional Molecular Components | |
|---|---|---|
| Kinetic control (catalysis) | Mineral surfaces Oxides: SiO2, TiO2, Al2O3 Hydroxides: Al(OH)3, Fe(OH)3 Iron sulfides: FeS, FeS2 Clays: montomorillonite, illite, bentonite, saponite, kaolinite Phosphates: hydroxyapatite | Amino acids Acid catalysis: His, Asp, Glu Basic catalysis: His, Arg, Lys Nucleophilic catalysis: Asp, Glu, Cys, Lys Di- and tripeptides Aldol reactions: Pro-based dipeptides (e.g., Pro-Gly, Pro-Phe, Pro-Val) Amide and phosphodiester bond formation: His-based di- and tripeptides (e.g., Ser-His, Ser-His-Gly, Ser-His-Asp) |
| Spatial control (compartmentalization) |  | |
| Energetic control (to favor endergonic reactions of interest) | Activating reagents | Activated monomers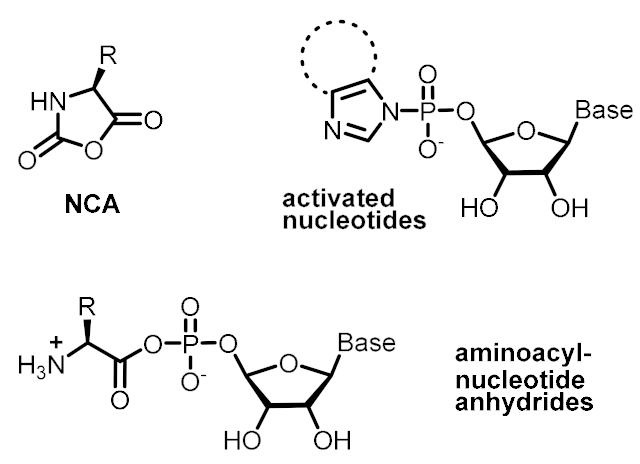 |
| Variability control (to achieve accurate recognition between functional components) | Canonical nucleobase pairing Alternative nucleobase pairing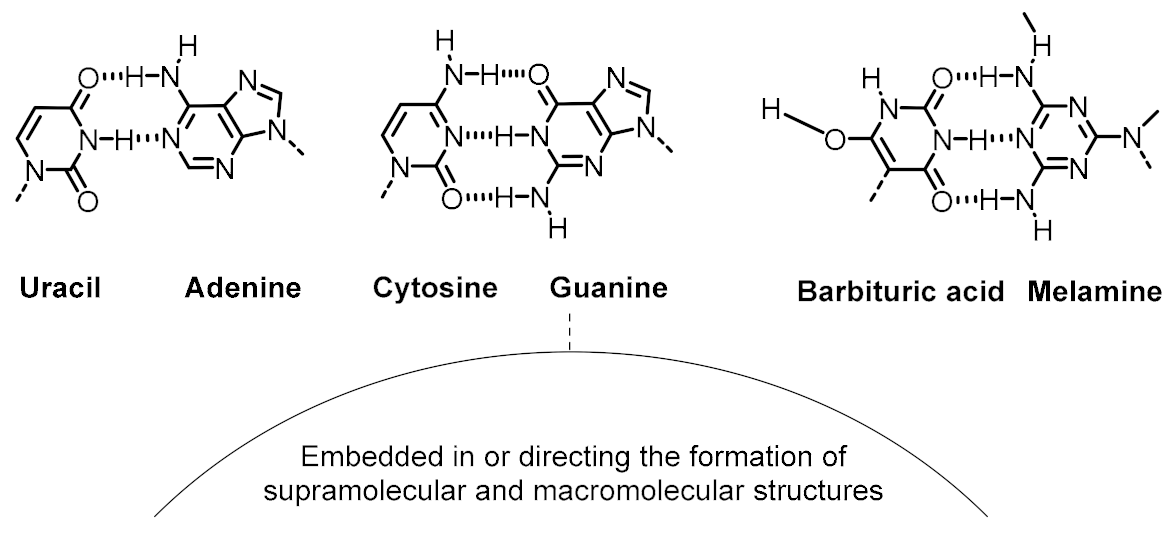 | |
| Control Mechanism | Functional Process |
|---|---|
| Entry 1: Autocatalytic reactions | 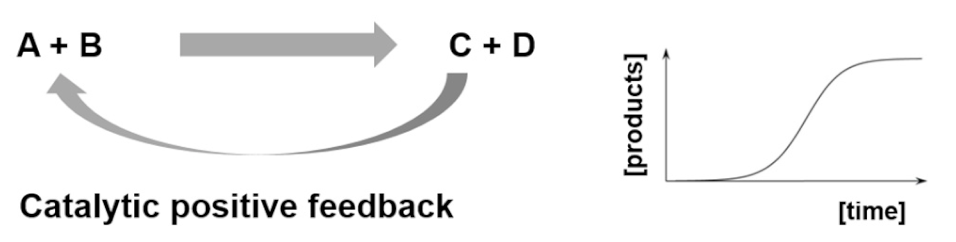 |
| Entry 2: Autocatalytic cycles |  |
| Entry 3: Stoichiometric couplings of autocatalytic cycles |  |
| Entry 4: Catalysis mediated autocatalytic sets |  |
| Entry 5: Catalysis in compartments | 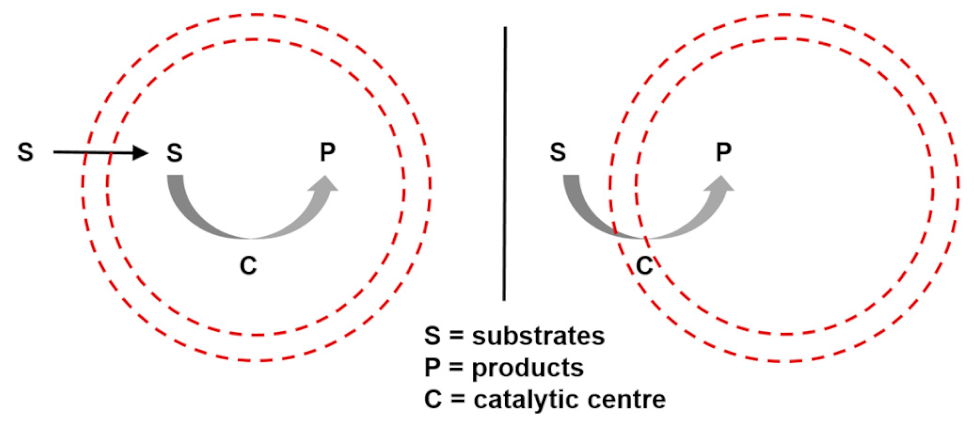 |
| Entry 6: Compartment self-reproduction |  |
| Entry 7: Osmotic couplings in compartments | 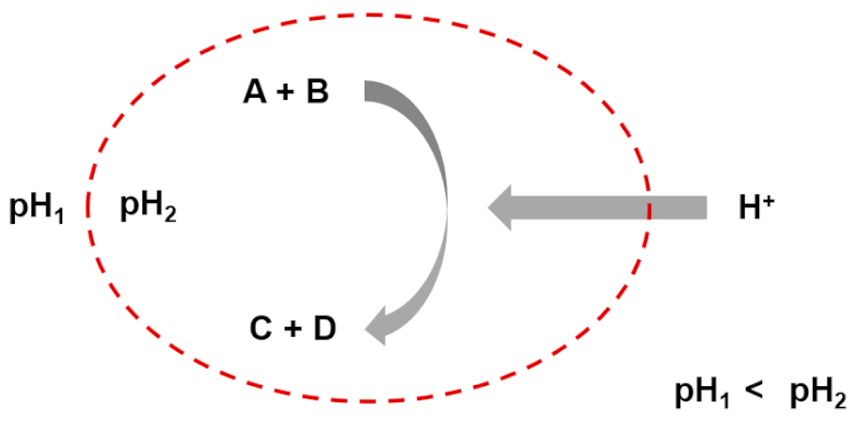 |
| Entry 8: Endergonic – exergonic couplings | 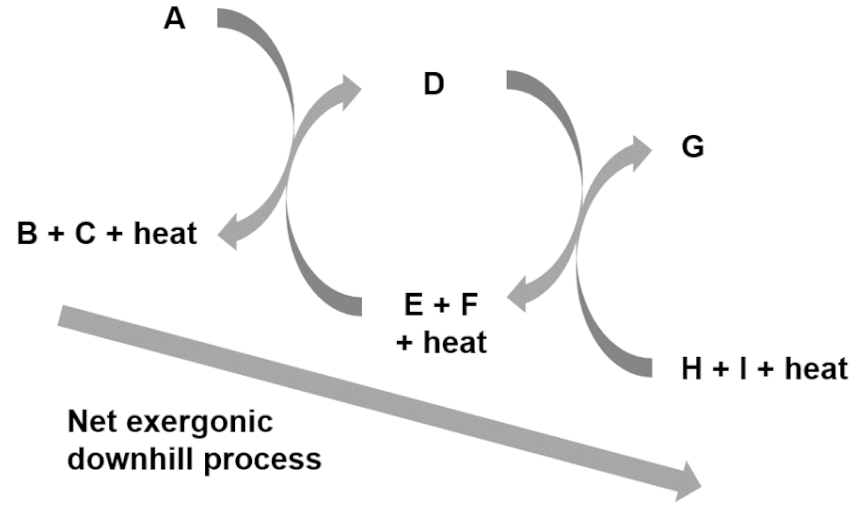 |
| Entry 9: Energy dissipation by self-assembly |  |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de la Escosura, A. The Informational Substrate of Chemical Evolution: Implications for Abiogenesis. Life 2019, 9, 66. https://doi.org/10.3390/life9030066
de la Escosura A. The Informational Substrate of Chemical Evolution: Implications for Abiogenesis. Life. 2019; 9(3):66. https://doi.org/10.3390/life9030066
Chicago/Turabian Stylede la Escosura, Andrés. 2019. "The Informational Substrate of Chemical Evolution: Implications for Abiogenesis" Life 9, no. 3: 66. https://doi.org/10.3390/life9030066
APA Stylede la Escosura, A. (2019). The Informational Substrate of Chemical Evolution: Implications for Abiogenesis. Life, 9(3), 66. https://doi.org/10.3390/life9030066





