Bioinformatic Analysis Reveals Archaeal tRNATyr and tRNATrp Identities in Bacteria
Abstract
:1. Introduction
2. Materials and Methods
Bioinformatics
3. Results
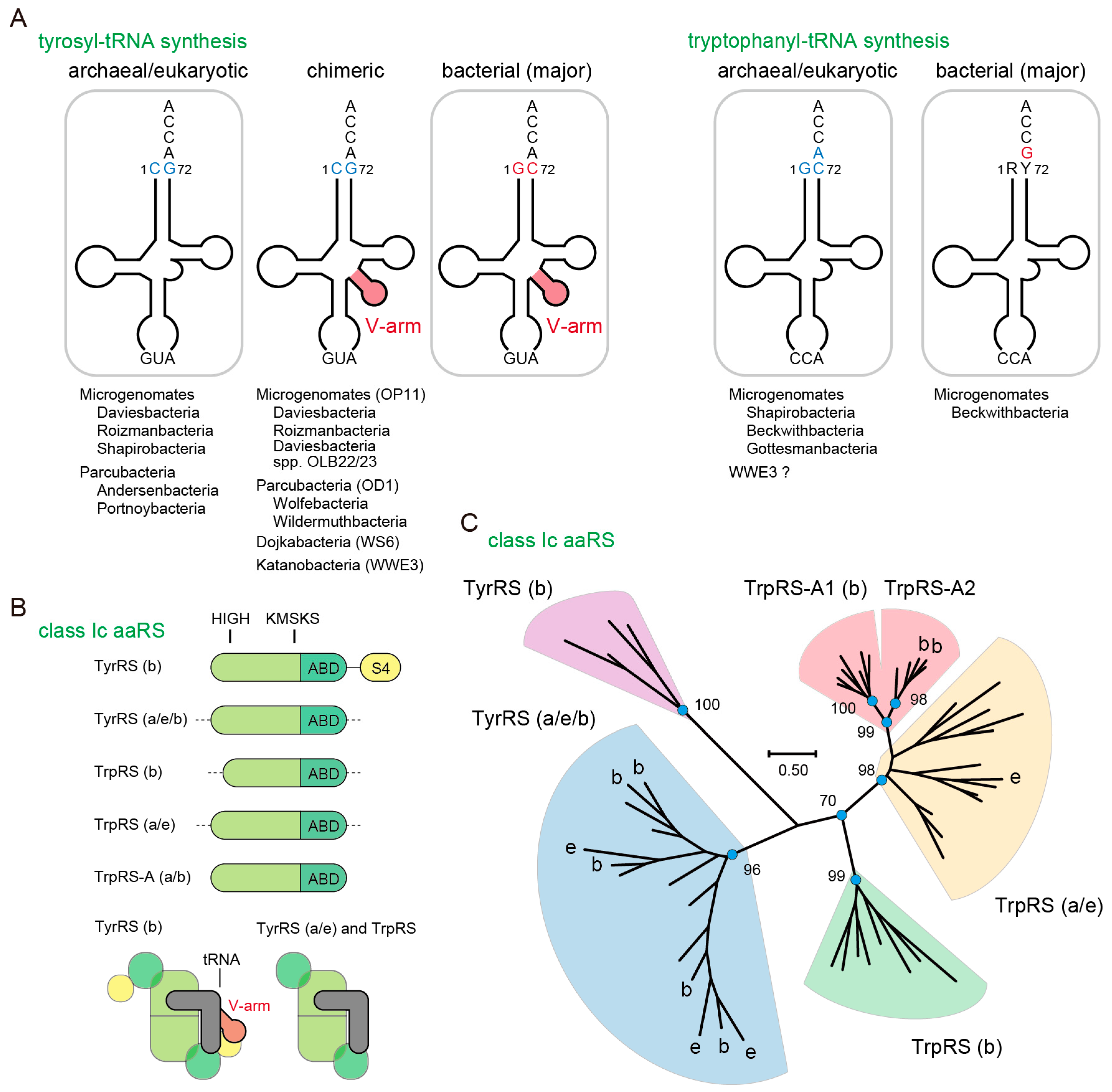
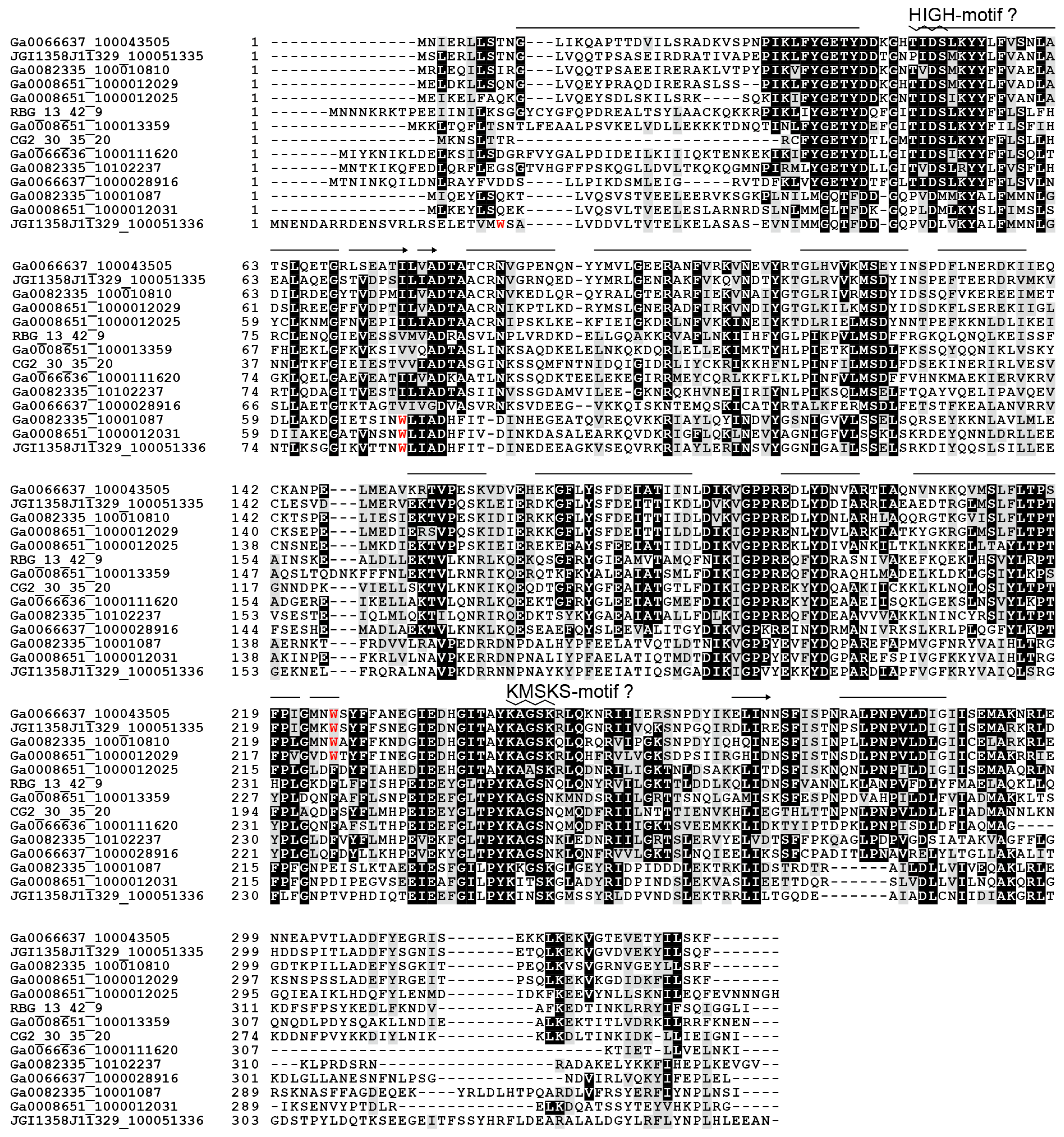
3.1. Identification of Non-Canonical Class Ic aaRS Sequences
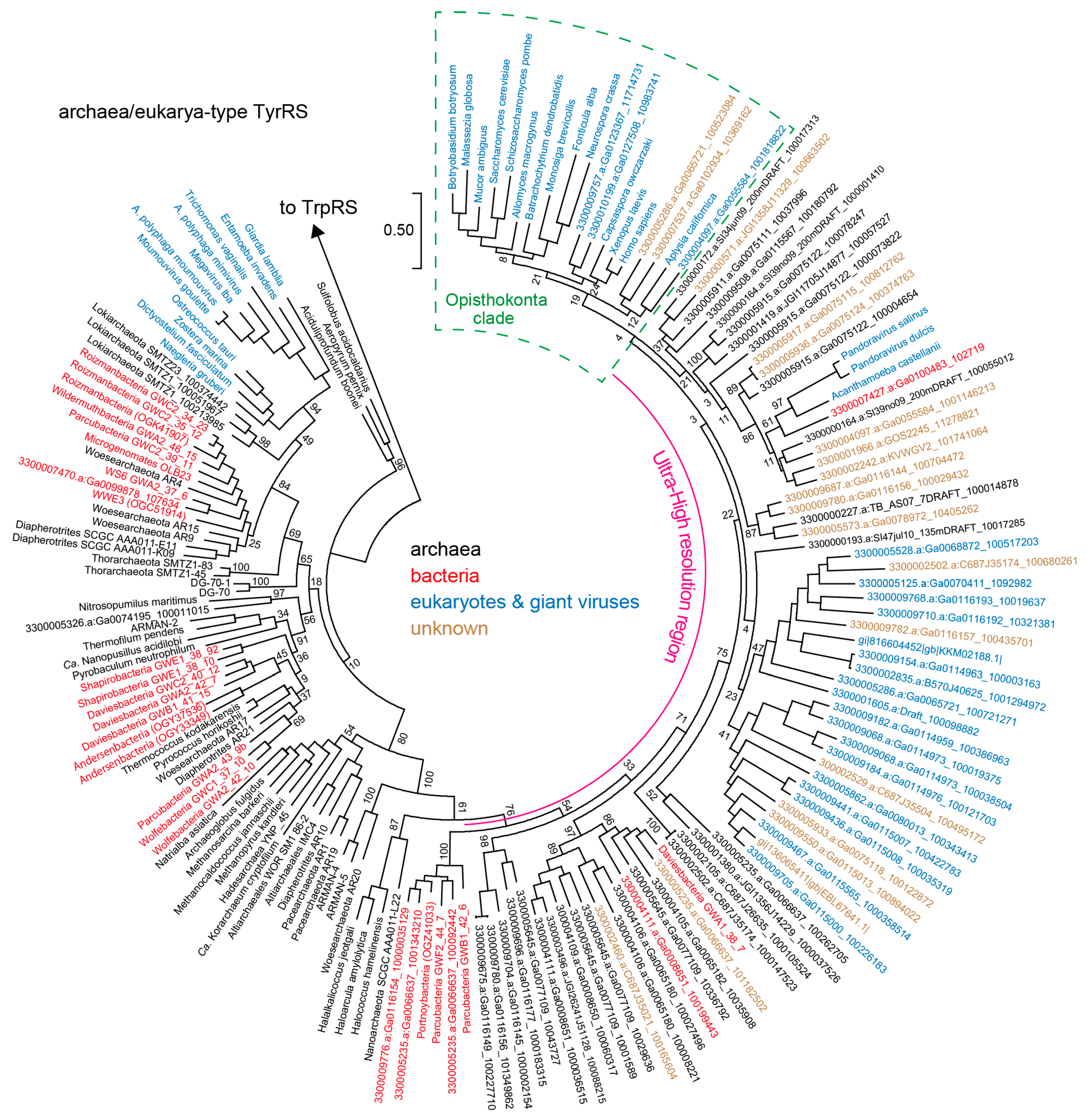
3.2. Archaea/Eukarya-Type TyrRS in the Bacterial Domain
3.3. Non-Canonical TrpRS Species and Their Associating Proteins
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CPR | candidate phyla radiation |
| aaRS | aminoacyl-tRNA synthetase |
| TyrRS | tyrosyl-tRNA synthetase |
| TrpRS | tryptophanyl-tRNA synthetase |
| PheRS | phenylalanyl-tRNA synthetase |
| HisRS | histidyl-tRNA synthetase |
| ORF | open reading frame |
| LGT | lateral gene transfer |
References
- Giegé, R.; Sissler, M.; Florentz, C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998, 26, 5017–5035. [Google Scholar] [CrossRef] [PubMed]
- Ibba, M.; Francklyn, C.; Cusack, S.E. The Aminoacyl-tRNA Synthetases; Landes Biosciences: Georgetown, TX, USA, 2005. [Google Scholar]
- Chaliotis, A.; Vlastaridis, P.; Mossialos, D.; Ibba, M.; Becker, H.D.; Stathopoulos, C.; Amoutzias, G.D. The complex evolutionary history of aminoacyl-tRNA synthetases. Nucleic Acids Res. 2016, 45, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.W., Jr. Tryptophanyl-tRNA synthetases. In The Aminoacyl-tRNA Synthetases; Ibba, M., Francklyn, C., Cusack, S.E., Eds.; Landes Biosciences: Georgetown, TX, USA, 2005. [Google Scholar]
- Bedouelle, H. Tyrosyl-tRNA synthetases. In The Aminoacyl-tRNA Synthetases; Ibba, M., Francklyn, C., Cusack, S.E., Eds.; Landes Biosciences: Georgetown, TX, USA, 2005. [Google Scholar]
- Bonnefond, L.; Giegé, R.; Rudinger-Thirion, J. Evolution of the tRNATyr/TyrRS aminoacylation systems. Biochimie 2005, 87, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Shen, W.; Giegé, R.; Wong, J.T. Identity elements of tRNATrp. Identification and evolutionary conservation. J. Biol. Chem. 1993, 268, 9316–9322. [Google Scholar] [PubMed]
- Furukawa, R.; Nakagawa, M.; Kuroyanagi, T.; Yokobori, S.I.; Yamagishi, A. Quest for Ancestors of Eukaryal Cells Based on Phylogenetic Analyses of Aminoacyl-tRNA Synthetases. J. Mol. Evol. 2017, 84, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Andam, C.P.; Gogarten, J.P. Biased gene transfer in microbial evolution. Nat. Rev. Microbiol. 2011, 9, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Shiba, K.; Motegi, H.; Schimmel, P. Maintaining genetic code through adaptations of tRNA synthetases to taxonomic domains. Trends Biochem. Sci. 1997, 22, 453–457. [Google Scholar] [CrossRef]
- Sassanfar, M.; Kranz, J.E.; Gallant, P.; Schimmel, P.; Shiba, K. A eubacterial Mycobacterium tuberculosis tRNA synthetase is eukaryote-like and resistant to a eubacterial-specific antisynthetase drug. Biochemistry 1996, 35, 9995–10003. [Google Scholar] [CrossRef] [PubMed]
- Ardell, D.H.; Andersson, S.G. TFAM detects co-evolution of tRNA identity rules with lateral transfer of histidyl-tRNA synthetase. Nucleic Acids Res. 2006, 34, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Fournier, G.P.; Alm, E.J. Ancestral Reconstruction of a Pre-LUCA Aminoacyl-tRNA Synthetase Ancestor Supports the Late Addition of Trp to the Genetic Code. J. Mol. Evol. 2015, 80, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Olsen, G.J.; Ibba, M.; Söll, D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000, 64, 202–236. [Google Scholar] [CrossRef] [PubMed]
- Wolf, Y.I.; Aravind, L.; Grishin, N.V.; Koonin, E.V. Evolution of aminoacyl-tRNA synthetases—Analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999, 9, 689–710. [Google Scholar] [PubMed]
- Ribas de Pouplana, L.; Frugier, M.; Quinn, C.L.; Schimmel, P. Evidence that two present-day components needed for the genetic code appeared after nucleated cells separated from eubacteria. Proc. Natl. Acad. Sci. USA 1996, 93, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Iraha, F.; Oki, K.; Kobayashi, T.; Ohno, S.; Yokogawa, T.; Nishikawa, K.; Yokoyama, S.; Sakamoto, K. Functional replacement of the endogenous tyrosyl-tRNA synthetase-tRNATyr pair by the archaeal tyrosine pair in Escherichia coli for genetic code expansion. Nucleic Acids Res. 2010, 38, 3682–3691. [Google Scholar] [CrossRef] [PubMed]
- Italia, J.S.; Addy, P.S.; Wrobel, C.J.J.; Crawford, L.A.; Lajoie, M.J.; Zheng, Y.; Chatterjee, A. An orthogonalized platform for genetic code expansion in both bacteria and eukaryotes. Nat. Chem. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Wolf, Y.I.; Koonin, E.V. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 2014, 466–467, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef] [PubMed]
- Eloe-Fadrosh, E.A.; Ivanova, N.N.; Woyke, T.; Kyrpides, N.C. Metagenomics uncovers gaps in amplicon-based detection of microbial diversity. Nat. Microbiol. 2016, 1, 15032. [Google Scholar] [CrossRef] [PubMed]
- Anantharaman, K.; Brown, C.T.; Hug, L.A.; Sharon, I.; Castelle, C.J.; Probst, A.J.; Thomas, B.C.; Singh, A.; Wilkins, M.J.; Karaoz, U. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 2016, 7, 13219. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, A.L.; Corel, E.; Pathmanathan, J.S.; Lopez, P.; Bapteste, E. Bipartite graph analyses reveal interdomain LGT involving ultrasmall prokaryotes and their divergent, membrane-related proteins. Environ. Microbiol. 2016, 18, 5072–5081. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.T.; Hug, L.A.; Thomas, B.C.; Sharon, I.; Castelle, C.J.; Singh, A.; Wilkins, M.J.; Wrighton, K.C.; Williams, K.H.; Banfield, J.F. Unusual biology across a group comprising more than 15% of domain bacteria. Nature 2015, 523, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Wrighton, K.C.; Castelle, C.J.; Varaljay, V.A.; Satagopan, S.; Brown, C.T.; Wilkins, M.J.; Thomas, B.C.; Sharon, I.; Williams, K.H.; Tabita, F.R.; et al. RubisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J. 2016, 10, 2702–2714. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, V.M.; Chen, I.M.; Chu, K.; Szeto, E.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Pagani, I.; Tringe, S.; et al. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 2014, 42, D568–D573. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Chen, X.L.; Guo, L.T.; Yu, Y.D.; Ding, J.P.; Jin, Y.X. Residues Lys-149 and Glu-153 switch the aminoacylation of tRNATrp in Bacillus subtilis. J. Biol. Chem. 2004, 279, 41960–41965. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Chen, X.; Xin, L.; Chen, L.; Jin, Y.; Wang, D. Species-specific differences in the operational RNA code for aminoacylation of tRNATrp. Nucleic Acids Res. 2001, 29, 4125–4133. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Otero, F.J.; Skene, R.J.; McRee, D.E.; Schimmel, P.; Ribas de Pouplana, L. Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains. Proc. Natl. Acad. Sci. USA 2003, 100, 15376–15380. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Nureki, O.; Ishitani, R.; Yaremchuk, A.; Tukalo, M.; Cusack, S.; Sakamoto, K.; Yokoyama, S. Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat. Struct. Biol. 2003, 10, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Stiebritz, M.T. A role for [Fe4S4] clusters in tRNA recognition—A theoretical study. Nucleic Acids Res. 2014, 42, 5426–5435. [Google Scholar] [CrossRef] [PubMed]
- Abergel, C.; Rudinger-Thirion, J.; Giegé, R.; Claverie, J.M. Virus-encoded aminoacyl-tRNA synthetases: Structural and functional characterization of mimivirus TyrRS and MetRS. J. Virol. 2007, 81, 12406–12417. [Google Scholar] [CrossRef] [PubMed]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Han, G.W.; Yang, X.L.; McMullan, D.; Chong, Y.E.; Krishna, S.S.; Rife, C.L.; Weekes, D.; Brittain, S.M.; Abdubek, P.; Ambing, E.; et al. Structure of a tryptophanyl-tRNA synthetase containing an iron-sulfur cluster. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 1326–1334. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Roy, H.; Ibba, M. Phenylalanyl-tRNA synthetase contains a dispensable RNA-binding domain that contributes to the editing of noncognate aminoacyl-tRNA. Biochemistry 2006, 45, 9156–9162. [Google Scholar] [CrossRef] [PubMed]
- Kapps, D.; Cela, M.; Théobald-Dietrich, A.; Hendrickson, T.; Frugier, M. OB or Not OB: Idiosyncratic utilization of the tRNA-binding OB-fold domain in unicellular, pathogenic eukaryotes. FEBS Lett. 2016, 590, 4180–4191. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.T.; Rossmann, M.G. Comparison of super-secondary structures in proteins. J. Mol. Biol. 1973, 76, 241–256. [Google Scholar] [CrossRef]
- Moutiez, M.; Belin, P.; Gondry, M. Aminoacyl-tRNA-Utilizing Enzymes in Natural Product Biosynthesis. Chem. Rev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Sobral, B.W.; Williams, K.P. Loss of a universal tRNA feature. J. Bacterial. 2007, 189, 1954–1962. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Gogakos, T.; Babina, A.M.; Söll, D.; Randau, L. Change of tRNA identity leads to a divergent orthogonal histidyl-tRNA synthetase/tRNAHis pair. Nucleic Acids Res. 2011, 39, 2286–2293. [Google Scholar] [CrossRef] [PubMed]
- Bonnefond, L.; Frugier, M.; Giegé, R.; Rudinger-Thirion, J. Human mitochondrial TyrRS disobeys the tyrosine identity rules. RNA 2005, 11, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Bonnefond, L.; Frugier, M.; Touzé, E.; Lorber, B.; Florentz, C.; Giegé, R.; Sauter, C.; Rudinger-Thirion, J. Crystal Structure of Human Mitochondrial Tyrosyl-tRNA Synthetase Reveals Common and Idiosyncratic Features. Structure 2007, 15, 1505–1516. [Google Scholar] [CrossRef] [PubMed]
- Buddha, M.R.; Crane, B.R. Structure and activity of an aminoacyl-tRNA synthetase that charges tRNA with nitro-tryptophan. Nat. Struct. Mol. Biol. 2005, 12, 274–275. [Google Scholar] [CrossRef] [PubMed]
- Kitabatake, M.; Ali, K.; Demain, A.; Sakamoto, K.; Yokoyama, S.; Söll, D. Indolmycin resistance of Streptomyces coelicolor A3(2) by induced expression of one of its two tryptophanyl-tRNA synthetases. J. Biol. Chem. 2002, 277, 23882–23887. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.Á.; Napolitano, M.; Ochoa de Alda, J.A.; Santamaría-Gómez, J.; Patterson, C.J.; Foster, A.W.; Bru-Martínez, R.; Robinson, N.J.; Luque, I. Trans-oligomerization of duplicated aminoacyl-tRNA synthetases maintains genetic code fidelity under stress. Nucleic Acids Res. 2015, 43, 9905–9917. [Google Scholar] [CrossRef] [PubMed]
- Buddha, M.R.; Keery, K.M.; Crane, B.R. An unusual tryptophanyl tRNA synthetase interacts with nitric oxide synthase in Deinococcus radiodurans. Proc. Natl. Acad. Sci. USA 2004, 101, 15881–15886. [Google Scholar] [CrossRef] [PubMed]
- Larson, E.T.; Kim, J.E.; Castaneda, L.J.; Napuli, A.J.; Zhang, Z.; Fan, E.; Zucker, F.H.; Verlinde, C.L.; Buckner, F.S.; Van Voorhis, W.C.; et al. The double-length tyrosyl-tRNA synthetase from the eukaryote Leishmania major forms an intrinsically asymmetric pseudo-dimer. J. Mol. Biol. 2011, 409, 159–176. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukai, T.; Reynolds, N.M.; Crnković, A.; Söll, D. Bioinformatic Analysis Reveals Archaeal tRNATyr and tRNATrp Identities in Bacteria. Life 2017, 7, 8. https://doi.org/10.3390/life7010008
Mukai T, Reynolds NM, Crnković A, Söll D. Bioinformatic Analysis Reveals Archaeal tRNATyr and tRNATrp Identities in Bacteria. Life. 2017; 7(1):8. https://doi.org/10.3390/life7010008
Chicago/Turabian StyleMukai, Takahito, Noah M. Reynolds, Ana Crnković, and Dieter Söll. 2017. "Bioinformatic Analysis Reveals Archaeal tRNATyr and tRNATrp Identities in Bacteria" Life 7, no. 1: 8. https://doi.org/10.3390/life7010008





