Toward Spatially Regulated Division of Protocells: Insights into the E. coli Min System from in Vitro Studies
Abstract
:1. Introduction
2. The E. coli Min System
3. Reconstitution of the Min System in Vitro
3.1. Self-Organization of Min Proteins into Surface Waves
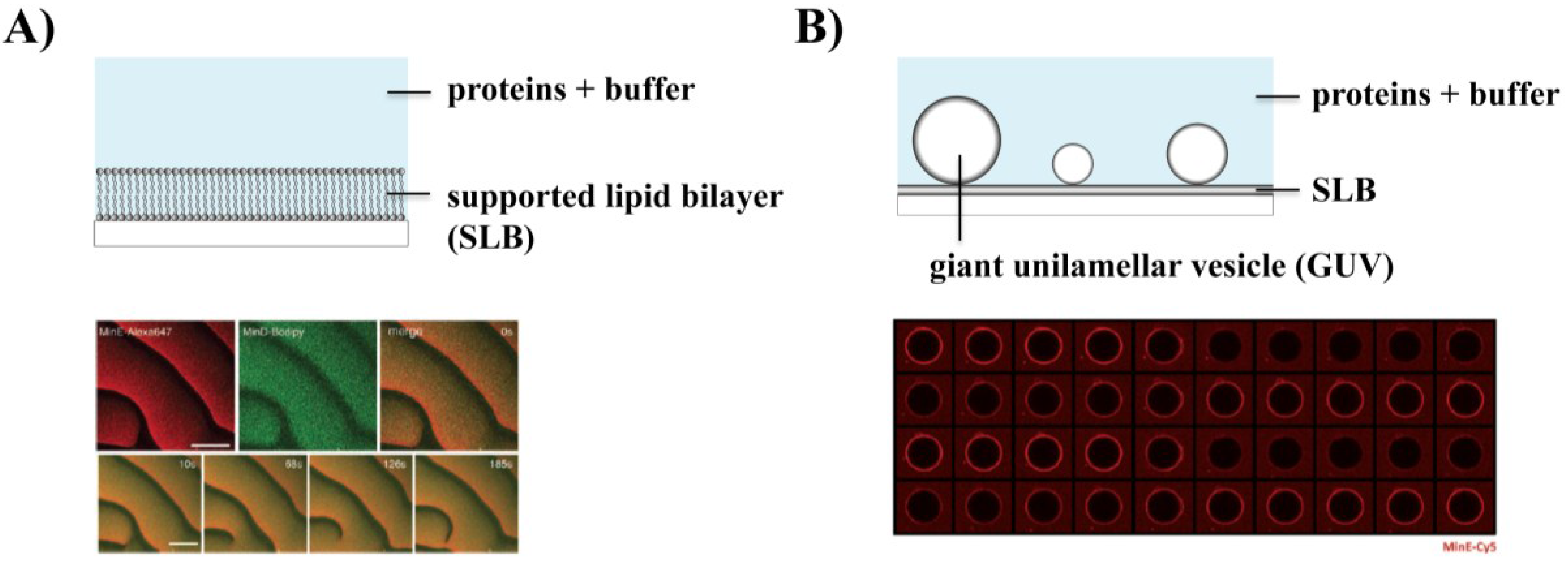
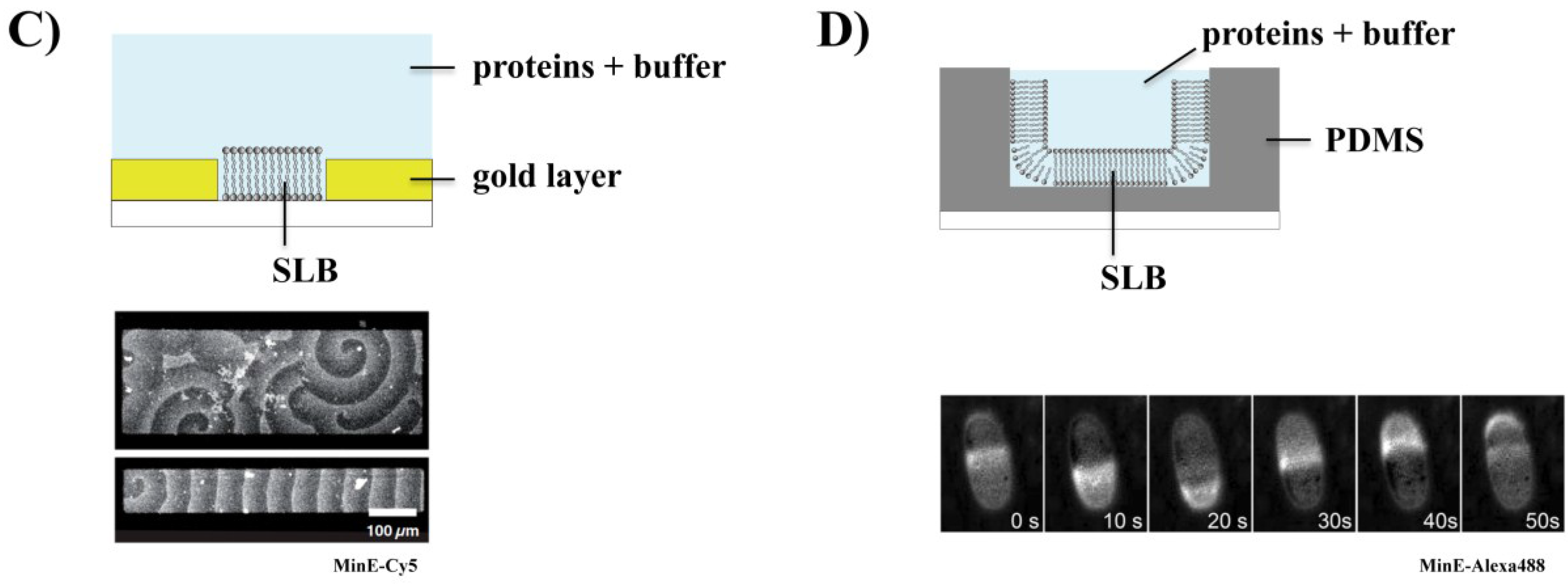
3.2. Effect of Physicochemical Parameters on Min Patterns
3.3. Effect of Geometry on Min Patterns: From Waves to Oscillations
3.4. Co-Reconstitution of the Min System with FtsZ
4. Conclusions and Outlook
Acknowledgments
Conflicts of Interest
References
- Luisi, P.L.; Ferri, F.; Stano, P. Approaches to semi-synthetic minimal cells: A review. Naturwissenschaften 2006, 93, 1–13. [Google Scholar]
- Dzieciol, A.J.; Mann, S. Designs for life: Protocell models in the laboratory. Chem. Soc. Rev. 2012, 41, 79–85. [Google Scholar]
- Mushegian, A.R.; Koonin, E.V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA 1996, 93, 10268–10273. [Google Scholar]
- Hutchison, C.A.; Peterson, S.N.; Gill, S.R.; Cline, R.T.; White, O.; Fraser, C.M.; Smith, H.O.; Venter, J.C. Global Transposon Mutagenesis and a Minimal Mycoplasma Genome. Science 1999, 286, 2165–2169. [Google Scholar]
- Schwille, P. Bottom-Up Synthetic Biology: Engineering in a Tinkerer’s World. Science 2011, 333, 1252–1254. [Google Scholar]
- Sole, R.V.; Munteanu, A.; Rodriguez-Caso, C.; Macia, J. Synthetic protocell biology: From reproduction to computation. Philos. Trans. R. Soc. B 2007, 362, 1727–1739. [Google Scholar]
- Chen, I.A.; Walde, P. From Self-Assembled Vesicles to Protocells. Cold Spring Harb. Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef]
- Noireaux, V.; Libchaber, A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl. Acad. Sci. USA 2004, 101, 17669–17674. [Google Scholar]
- Rasmussen, S.; Chen, L.; Nilsson, M.; Abe, S. Bridging nonliving and living matter. Artif. Life 2003, 9, 269–316. [Google Scholar]
- Martos, A.; Jimenez, M.; Rivas, G.; Schwille, P. Towards a bottom-up reconstitution of bacterial cell division. Trends Cell Biol. 2012, 22, 634–643. [Google Scholar]
- Osawa, M.; Anderson, D.E.; Erickson, H.P. Reconstitution of Contractile FtsZ Rings in Liposomes. Science 2008, 320, 792–794. [Google Scholar]
- Vicente, M.; Rico, A.I. The order of the ring: Assembly of Escherichia coli cell division components. Mol. Microbiol. 2006, 61, 5–8. [Google Scholar]
- Lutkenhaus, J.; Pichoff, S.; Du, S. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton 2012, 69, 778–790. [Google Scholar]
- Wu, L.J.; Errington, J. Nucleoid occlusion and bacterial cell division. Nat. Rev. Microbiol. 2012, 10, 8–12. [Google Scholar]
- Bernhardt, T.G.; de Boer, P.A. SlmA, a Nucleoid-Associated, FtsZ Binding Protein Required for Blocking Septal Ring Assembly over Chromosomes in E. coli. Mol. Cell 2005, 18, 555–564. [Google Scholar]
- Dajkovic, A.; Lan, G.; Sun, S.X.; Wirtz, D.; Lutkenhaus, J. MinC Spatially Controls Bacterial Cytokinesis by Antagonizing the Scaffolding Function of FtsZ. Curr. Biol. 2008, 18, 235–244. [Google Scholar]
- De Boer, P.A.; Crossley, R.E.; Hand, A.R.; Rothfield, L.I. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 1991, 10, 4371–4380. [Google Scholar]
- Hu, Z.; Mukherjee, A.; Pichoff, S.; Lutkenhaus, J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 1999, 96, 14819–14824. [Google Scholar]
- Hu, Z.; Lutkenhaus, J. Topological Regulation of Cell Division in E. coli: Spatiotemporal Oscillation of MinD Requires Stimulation of Its ATPase by MinE and Phospholipid. Mol. Cell 2001, 7, 1337–1343. [Google Scholar]
- Hu, Z.; Lutkenhaus, J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 1999, 34, 82–90. [Google Scholar]
- Raskin, D.M.; de Boer, P.A. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 1999, 96, 4971–4976. [Google Scholar]
- Adler, H.I.; Fisher, W.D.; Cohen, A.; Hardigree, A.A. Miniature Escherichia coli cells deficient in DNA. Proc. Natl. Acad. Sci. USA 1967, 57, 321–326. [Google Scholar]
- Loose, M.; Kruse, K.; Schwille, P. Protein Self-Organization: Lessons from the Min System. Annu. Rev. Biophys. 2011, 40, 315–336. [Google Scholar]
- Lutkenhaus, J. Min oscillation in bacteria. Adv. Exp. Med. Biol. 2008, 641, 49–61. [Google Scholar]
- Shih, Y.L.; Zheng, M. Spatial control of the cell division site by the Min system in Escherichia coli. Environ. Microbiol. 2013, 15, 3229–3239. [Google Scholar]
- Hu, Z.; Lutkenhaus, J. Analysis of MinC Reveals Two Independent Domains Involved in Interaction with MinD and FtsZ. J. Bacteriol. 2000, 182, 3965–3971. [Google Scholar]
- Cordell, S.C.; Anderson, R.E.; Lowe, J. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 2001, 20, 2454–2461. [Google Scholar]
- Johnson, J.E.; Lackner, L.L.; de Boer, P.A. Targeting of DMinC/MinD and DMinC/DicB Complexes to Septal Rings in Escherichia coli Suggests a Multistep Mechanism for MinC-Mediated Destruction of Nascent FtsZ Rings. J. Bacteriol. 2002, 184, 2951–2962. [Google Scholar]
- Shen, B.; Lutkenhaus, J. Examination of the interaction between FtsZ and MinCN in E. coli suggests how MinC disrupts Z rings. Mol. Microbiol. 2010, 75, 1285–1298. [Google Scholar]
- Raskin, D.M.; de Boer, P.A. MinDE-Dependent Pole-to-Pole Oscillation of Division Inhibitor MinC in Escherichia coli. J. Bacteriol. 1999, 181, 6419–6424. [Google Scholar]
- Lutkenhaus, J.; Sundaramoorthy, M. MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol. Microbiol. 2003, 48, 295–303. [Google Scholar]
- Lackner, L.L.; Raskin, D.M.; de Boer, P.A.J. ATP-Dependent Interactions between Escherichia coli Min Proteins and the Phospholipid Membrane in Vitro. J. Bacteriol. 2003, 185, 735–749. [Google Scholar]
- Szeto, T.H.; Rowland, S.L.; Habrukowich, C.L.; King, G.F. The MinD Membrane Targeting Sequence is a Transplantable Lipid-Binding Helix. J. Biol. Chem. 2003, 278, 40050–40056. [Google Scholar]
- Szeto, T.H.; Rowland, S.L.; Rothfield, L.I.; King, G.F. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc. Natl. Acad. Sci. USA 2002, 99, 15693–15698. [Google Scholar]
- Hu, Z.; Lutkenhaus, J. A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol. Microbiol. 2003, 47, 345–355. [Google Scholar]
- Renner, L.D.; Weibel, D.B. MinD and MinE Interact with Anionic Phospholipids and Regulate Division Plane Formation in Escherichia coli. J. Biol. Chem. 2012, 287, 38835–38844. [Google Scholar]
- Ma, L.; King, G.F.; Rothfield, L. Positioning of the MinE binding site on the MinD surface suggests a plausible mechanism for activation of the Escherichia coli MinD ATPase during division site selection. Mol. Microbiol. 2004, 54, 99–108. [Google Scholar]
- Ma, L.Y.; King, G.; Rothfield, L. Mapping the MinE Site Involved in Interaction with the MinD Division Site Selection Protein of Escherichia coli. J. Bacteriol. 2003, 185, 4948–4955. [Google Scholar]
- Pichoff, S.; Vollrath, B.; Touriol, C.; Bouche, J.P. Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol. Microbiol. 1995, 18, 321–329. [Google Scholar]
- Zhao, C.R.; de Boer, P.A.; Rothfield, L.I. Proper placement of the Escherichia coli division site requires two functions that are associated with different domains of the MinE protein. Proc. Natl. Acad. Sci. USA 1995, 92, 4313–4317. [Google Scholar]
- Wu, W.; Park, K.T.; Holyoak, T.; Lutkenhaus, J. Determination of the structure of the MinD-ATP complex reveals the orientation of MinD on the membrane and the relative location of the binding sites for MinE and MinC. Mol. Microbiol. 2011, 79, 1515–1528. [Google Scholar]
- Hsieh, C.W.; Lin, T.Y.; Lai, H.M.; Lin, C.C.; Hsieh, T.S.; Shih, Y.L. Direct MinE-membrane interaction contributes to the proper localization of MinDE in E. coli. Mol. Microbiol. 2010, 75, 499–512. [Google Scholar]
- Park, K.T.; Wu, W.; Battaile, K.P.; Lovell, S.; Holyoak, T.; Lutkenhaus, J. The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis. Cell 2011, 146, 396–407. [Google Scholar]
- Ghasriani, H.; Ducat, T.; Hart, C.T.; Hafizi, F.; Chang, N.; Al-Baldawi, A.; Ayed, S.H.; Lundstrom, P.; Dillon, J.A.; Goto, N.K.; et al. Appropriation of the MinD protein-interaction motif by the dimeric interface of the bacterial cell division regulator MinE. Proc. Natl. Acad. Sci. USA 2010, 107, 18416–18421. [Google Scholar]
- Kang, G.B.; Song, H.E.; Kim, M.K.; Youn, H.S.; Lee, J.G.; An, J.Y.; Chun, J.S.; Jeon, H.; Eom, S.H. Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor. Mol. Microbiol. 2010, 76, 1222–1231. [Google Scholar]
- King, G.F.; Shih, Y.L.; Maciejewski, M.W.; Bains, N.P.; Pan, B.; Rowland, S.L.; Mullen, G.P.; Rothfield, L.I. Structural basis for the topological specificity function of MinE. Nat. Struct. Biol. 2000, 7, 1013–1017. [Google Scholar]
- Zheng, M.; Chiang, Y.L.; Lee, H.L.; Kong, L.R.; Hsu, S.T.; Hwang, I.S.; Rothfield, L.I.; Shih, Y.L. Self-Assembly of MinE on the Membrane Underlies Formation of the MinE Ring to Sustain Function of the Escherichia coli Min System. J. Biol. Chem. 2014, 289, 21252–21266. [Google Scholar]
- Hale, C.A.; Meinhardt, H.; de Boer, P.A. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 2001, 20, 1563–1572. [Google Scholar]
- Raskin, D.M.; de Boer, P.A. The MinE Ring: An FtsZ-Independent Cell Structure Required for Selection of the Correct Division Site in E. coli. Cell 1997, 91, 685–694. [Google Scholar]
- Jia, S.; Keilberg, D.; Hot, E.; Thanbichler, M.; Sogaard-Andersen, L.; Lenz, P. Effect of the Min System on Timing of Cell Division in Escherichia coli. PLoS One 2014, 9. [Google Scholar] [CrossRef]
- Di Ventura, B.; Knecht, B.; Andreas, H.; Godinez, W.J.; Fritsche, M.; Rohr, K.; Nickel, W.; Heermann, D.W.; Sourjik, V. Chromosome segregation by the Escherichia coli Min system. Mol. Syst. Biol. 2013, 9. [Google Scholar] [CrossRef]
- Loose, M.; Fischer-Friedrich, E.; Ries, J.; Kruse, K.; Schwille, P. Spatial Regulators for Bacterial Cell Division Self-Organize into Surface Waves in Vitro. Science 2008, 320, 789–792. [Google Scholar]
- Camazine, S. Self-Organization in Biological Systems; Princeton University Press: Princeton, NJ, USA, 2001. [Google Scholar]
- Nettesheim, S.; Vonoertzen, A.; Rotermund, H.H.; Ertl, G. Reaction-diffusion patterns in the catalytic CO oxidation on Pt(110): Front propagation and spiral waves. J. Chem. Phys. 1993, 98, 9977–9985. [Google Scholar]
- Sagues, F.; Epstein, I.R. Nonlinear chemical dynamics. Dalton Trans. 2003. [Google Scholar] [CrossRef]
- Martos, A.; Petrasek, Z.; Schwille, P. Propagation of MinCDE waves on free-standing membranes. Environ. Microbiol. 2013, 15, 3319–3326. [Google Scholar]
- Schweizer, J.; Loose, M.; Bonny, M.; Kruse, K.; Monch, I.; Schwille, P. Geometry sensing by self-organized protein patterns. Proc. Natl. Acad. Sci. USA 2012, 109, 15283–15288. [Google Scholar]
- Zieske, K.; Schwille, P. Reconstitution of Pole-to-Pole Oscillations of Min proteins in Microengineered Polydimethylsiloxane Compartments. Angew. Chem. 2013, 52, 459–462. [Google Scholar]
- Meacci, G.; Kruse, K. Min-oscillations in Escherichia coli induced by interactions of membrane-bound proteins. Phys. Biol. 2005, 2, 89–97. [Google Scholar]
- Loose, M.; Fischer-Friedrich, E.; Herold, C.; Kruse, K.; Schwille, P. Min protein patterns emerge from rapid rebinding and membrane interaction of MinE. Nat. Struct. Mol. Biol. 2011, 18, 577–583. [Google Scholar]
- Przybylo, M.; Sykora, J.; Humpolickova, J.; Benda, A.; Zan, A.; Hof, M. Lipid Diffusion in Giant Unilamellar Vesicles is More than 2 Times Faster than in Supported Phospholipid Bilayers under Identical Conditions. Langmuir 2006, 22, 9096–9099. [Google Scholar]
- Ivanov, V.; Mizuuchi, K. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 8071–8078. [Google Scholar]
- Vecchiarelli, A.G.; Li, M.; Mizuuchi, M.; Mizuuchi, K. Differential affinities of MinD and MinE to anionic phospholipid influence Min patterning dynamics in vitro. Mol. Microbiol. 2014, 93, 453–463. [Google Scholar]
- Corbin, B.D.; Yu, X.C.; Margolin, W. Exploring intracellular space: Function of the Min system in round-shaped Escherichia coli. EMBO J. 2002, 21, 1998–2008. [Google Scholar]
- Fu, X.; Shih, Y.L.; Zhang, Y.; Rothfield, L.I. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 2001, 98, 980–985. [Google Scholar]
- Zieske, K.; Schweizer, J.; Schwille, P. Surface topology assisted alignment of Min protein waves. FEBS Lett. 2014, 588, 2545–2549. [Google Scholar]
- Rivas, G.; Vogel, S.K.; Schwille, P. Reconstitution of cytoskeletal protein assemblies for large-scale membrane transformation. Curr. Opin. Chem. Biol. 2014, 22, 18–26. [Google Scholar]
- Osawa, M.; Erickson, H.P. Liposome division by a simple bacterial division machinery. Proc. Natl. Acad. Sci. USA 2013, 110, 11000–11004. [Google Scholar]
- Loose, M.; Mitchison, T.J. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 2014, 16, 38–46. [Google Scholar]
- Schwille, P. Bacterial Cell Division: A Swirling Ring to Rule Them All? Curr. Biol. 2014, 24, R157–R159. [Google Scholar]
- Arumugam, S.; Petrasek, Z.; Schwille, P. MinCDE exploits the dynamic nature of FtsZ filaments for its spatial regulation. Proc. Natl. Acad. Sci. USA 2014, 111, E1192–E1200. [Google Scholar]
- Zieske, K.; Schwille, P. Reconstitution of self-organizing protein gradients as spatial cues in cell-free systems. eLife 2014, 3. [Google Scholar] [CrossRef]
- Arumugam, S.; Chwastek, G.; Fischer-Friedrich, E.; Ehrig, C.; Monch, I.; Schwille, P. Surface Topology Engineering of Membranes for the Mechanical Investigation of the Tubulin Homologue FtsZ. Angew. Chem. 2012, 51, 11858–11862. [Google Scholar]
- Van Swaay, D.; de Mello, A. Microfluidic methods for forming liposomes. Lab Chip 2013, 13, 752–767. [Google Scholar]
- Pautot, S.; Frisken, B.J.; Weitz, D.A. Engineering asymmetric vesicles. Proc. Natl. Acad. Sci. USA 2003, 100, 10718–10721. [Google Scholar]
- Stachowiak, J.C.; Richmond, D.L.; Li, T.H.; Liu, A.P.; Parekh, S.H.; Fletcher, D.A. Unilamellar vesicle formation and encapsulation by microfluidic jetting. Proc. Natl. Acad. Sci. USA 2008, 105, 4697–4702. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kretschmer, S.; Schwille, P. Toward Spatially Regulated Division of Protocells: Insights into the E. coli Min System from in Vitro Studies. Life 2014, 4, 915-928. https://doi.org/10.3390/life4040915
Kretschmer S, Schwille P. Toward Spatially Regulated Division of Protocells: Insights into the E. coli Min System from in Vitro Studies. Life. 2014; 4(4):915-928. https://doi.org/10.3390/life4040915
Chicago/Turabian StyleKretschmer, Simon, and Petra Schwille. 2014. "Toward Spatially Regulated Division of Protocells: Insights into the E. coli Min System from in Vitro Studies" Life 4, no. 4: 915-928. https://doi.org/10.3390/life4040915




