A Survey of Protein Structures from Archaeal Viruses
Abstract
:1. Introduction
| Virus | Virus Family | Host |
|---|---|---|
| Haloarcula hispanica virus (SH1) | unclassified | Haloarcula hispanica, Halorubrum, Haloferax [12,13] |
| Sulfolobus turreted icosahedral virus (STIV) | unclassified | Sulfolobus [14] |
| Acidianus filamentous virus 1 (AFV1) | Lipothrixviridae | Acidianus [15] |
| Sulfolobus islandicus rod-shaped virus 1 (SIRV1) | Rudiviridae | Sulfolobus [16] |
| Sulfolobus spindle-shaped virus 1 (SSV1) | Fuselloviridae | Sulfolobus [17] |
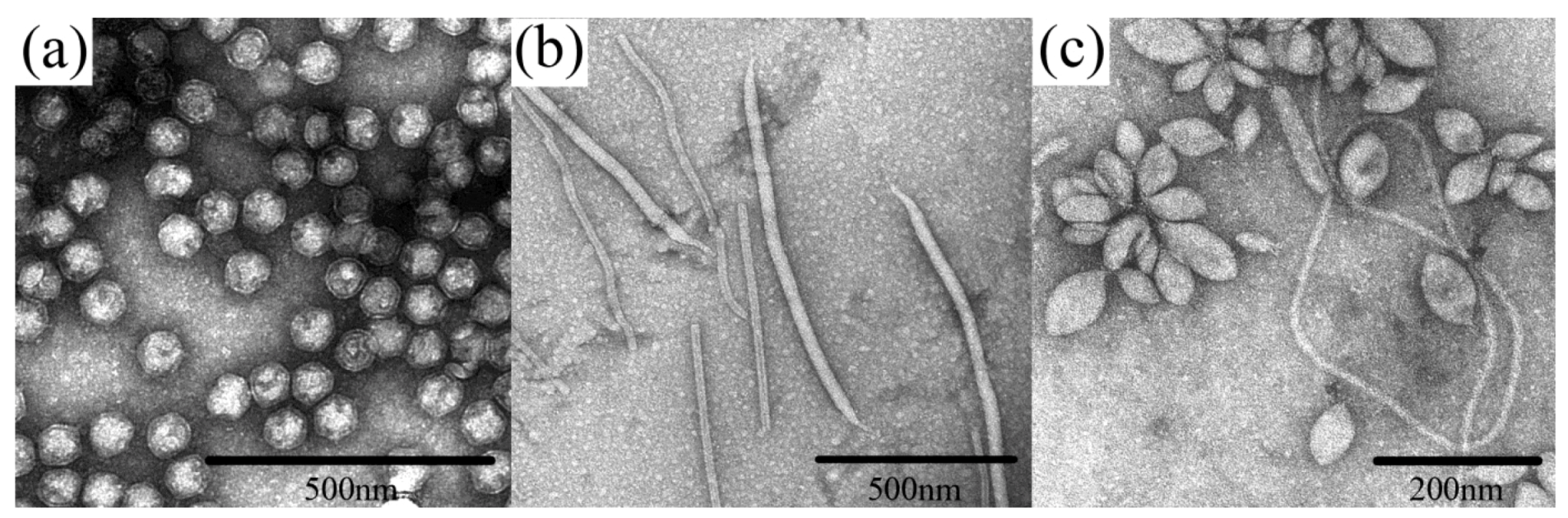
2. Discussion
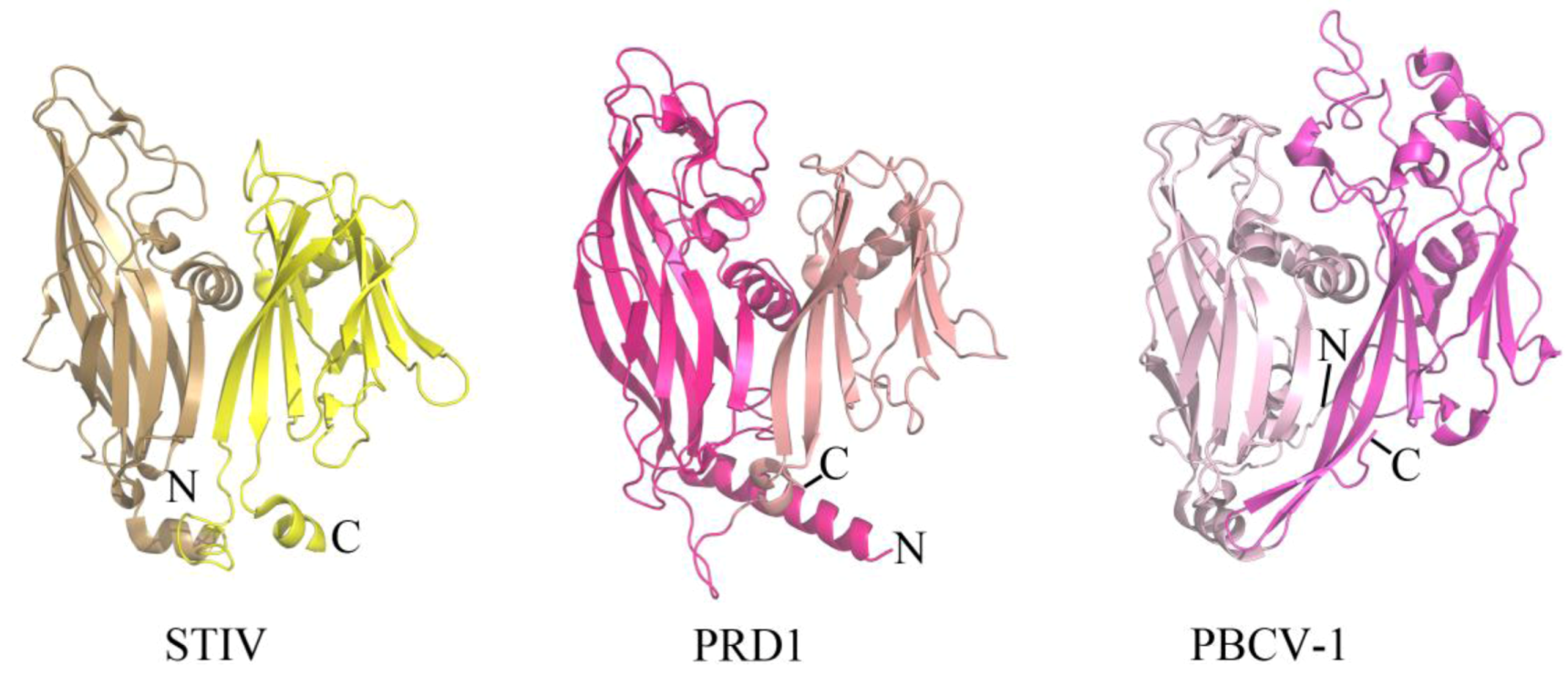
2.1. Evolutionary Links Revealed Through Structures of Archaeal Virus Major Capsid Proteins
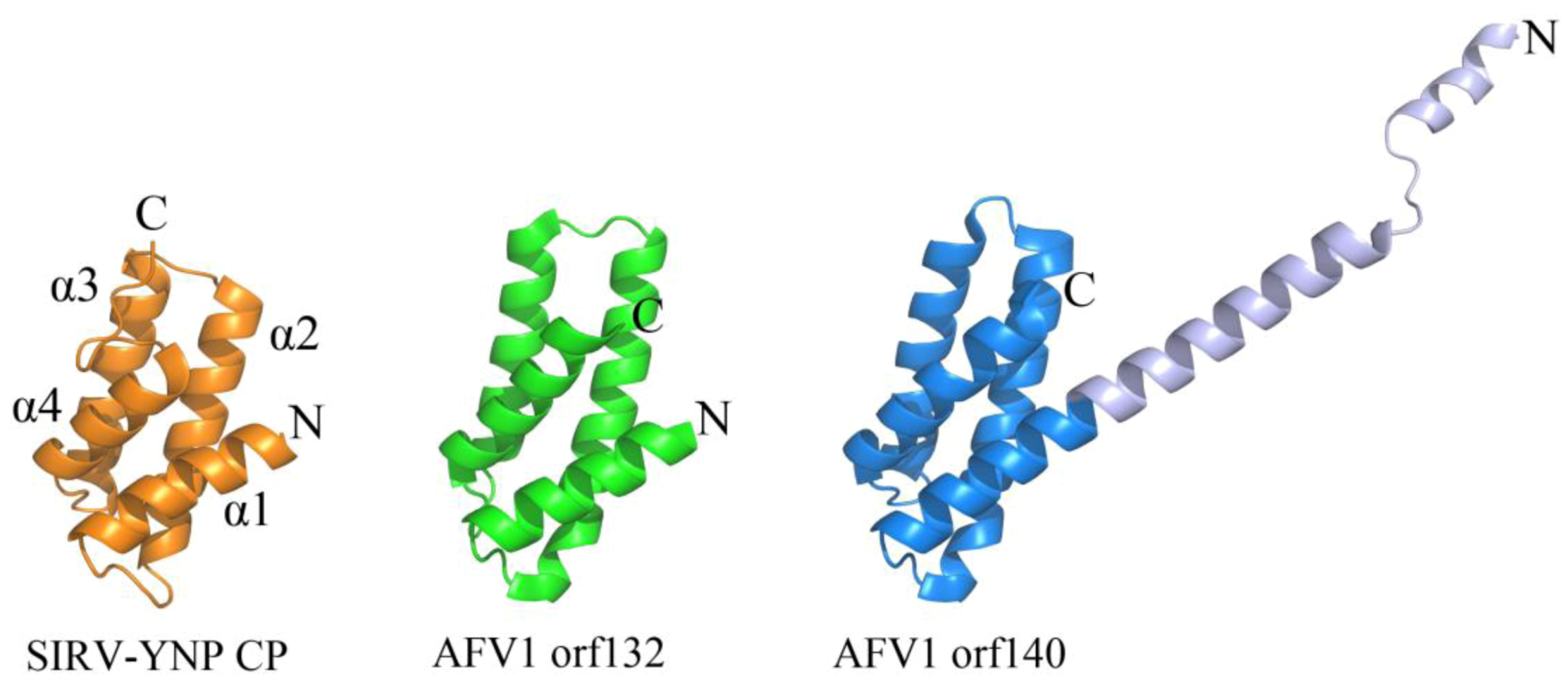
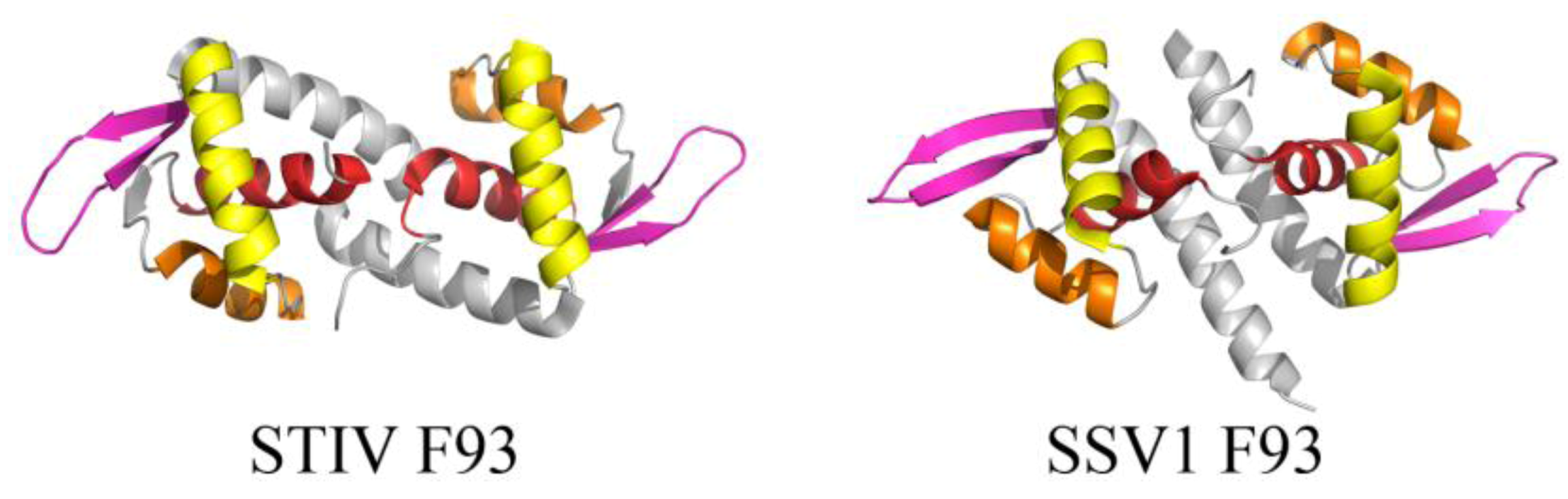
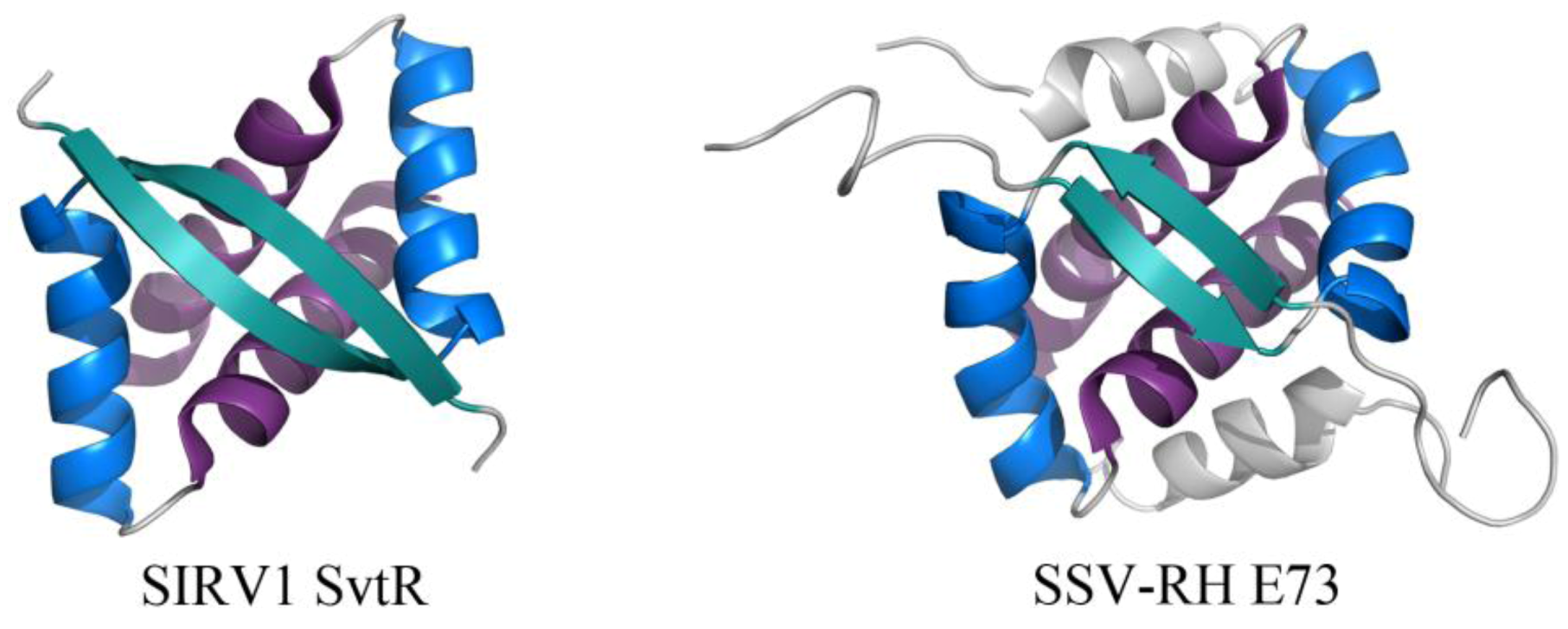
2.2. Common DNA-Binding Motifs Observed in Proteins from Archaeal Viruses
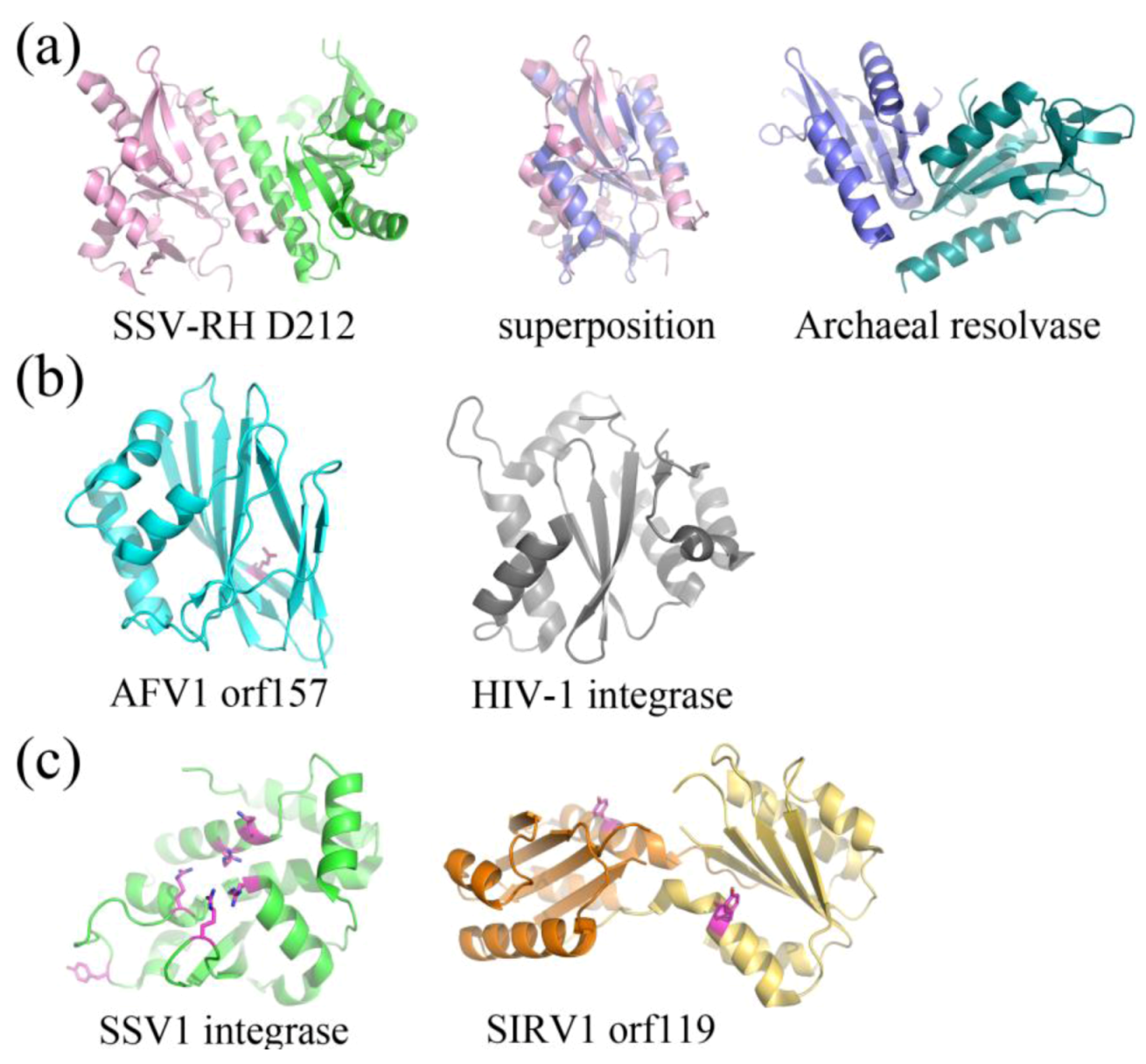
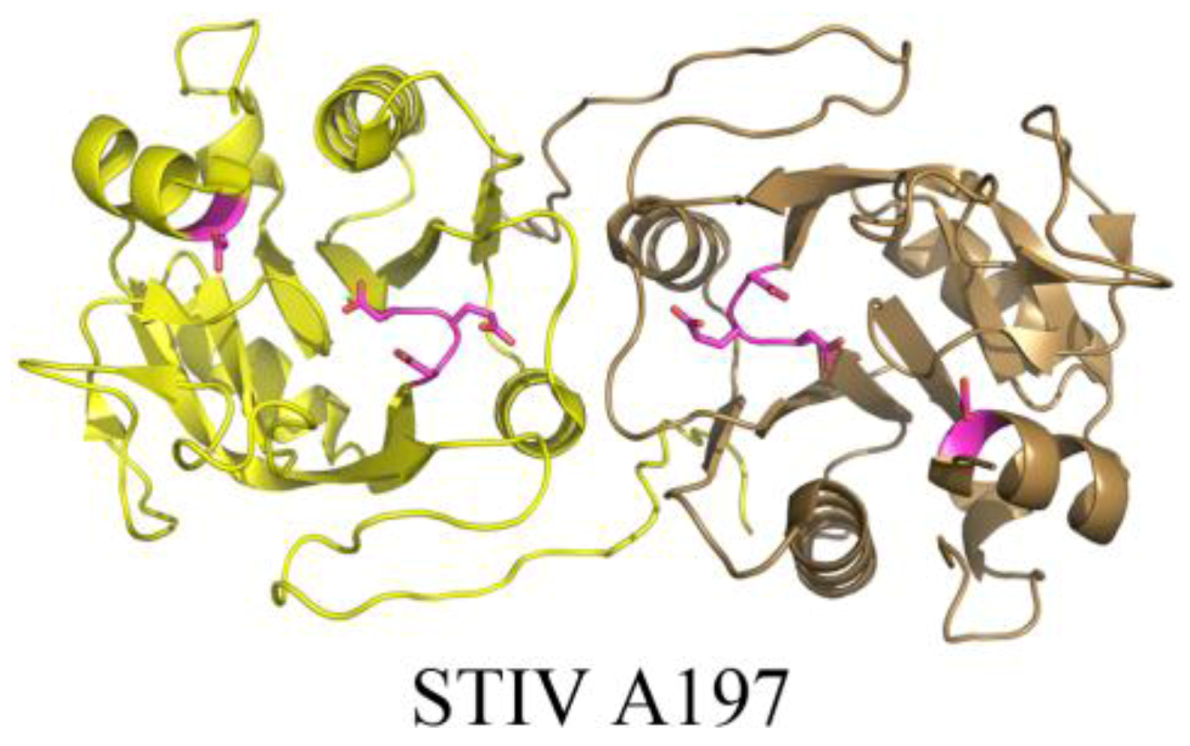
2.3. Structurally Characterized Enzymes of Archaeal Viruses
3. Conclusions
Acknowledgments
References
- Meile, L.; Jenal, U.; Studer, D.; Jordan, M.; Leisinger, T. Characterization of psi-m1, a virulent phage of methanobacterium-thermoautotrophicum marburg. Arch. Microbiol. 1989, 152, 105–110. [Google Scholar] [CrossRef]
- Pfister, P.; Wesserfallen, A.; Stettler, R.; Leisinger, T. Molecular analysis of methanobacterium phage psi m2. Mol. Microbiol. 1998, 30, 233–244. [Google Scholar] [CrossRef]
- Brochier-Armanet, C.; Boussau, B.; Gribaldo, S.; Forterre, P. Mesophilic crenarchaeota: Proposal for a third archaeal phylum, the thaumarchaeota. Nat. Rev. Microbiol. 2008, 6, 245–252. [Google Scholar] [CrossRef]
- Williamson, M. How Proteins Work; Garland Publishing Inc: New York, NY, 2011; p. 464. [Google Scholar]
- Lawrence, C.M.; Menon, S.; Eilers, B.J.; Bothner, B.; Khayat, R.; Douglas, T.; Young, M.J. Structural and functional studies of archaeal viruses. J. Biol. Chem. 2009, 284, 12599–12603. [Google Scholar]
- Prangishvili, D. Evolutionary insights from studies on viruses of hyperthermophilic archaea. Res. Microbiol. 2003, 154, 289–294. [Google Scholar]
- Le Romancer, M.; Gaillard, M.; Geslin, C.; Prieur, D. Viruses in extreme environments. Rev. Environ. Sci. Biotechnol. 2007, 6, 17–31. [Google Scholar] [CrossRef] [Green Version]
- Prangishvill, D.; Garrett, R.A.; Koonin, E.V. Evolutionary genomics of archaeal viruses: Unique viral genomes in the third domain of life. Virus Res. 2006, 117, 52–67. [Google Scholar] [CrossRef]
- Prangishvili, D.; Forterre, P.; Garrett, R.A. Viruses of the archaea: A unifying view. Nat. Rev. Microbiol. 2006, 4, 837–848. [Google Scholar] [CrossRef]
- Jaalinoja, H.T.; Roine, E.; Laurinmaki, P.; Kivela, H.M.; Bamford, D.H.; Butcher, S.J. Structure and host-cell interaction of sh1, a membrane-containing, halophilic euryarchaeal virus. Proc. Natl. Acad. Sci. USA 2008, 105, 8008–8013. [Google Scholar]
- Pietila, M.K.; Laurinmaki, P.; Russell, D.A.; Ko, C.C.; Jacobs-Sera, D.; Butcher, S.J.; Bamford, D.H.; Hendrix, R.W. Insights into head-tailed viruses infecting extremely halophilic archaea. J. Virol. 2013. [Google Scholar]
- Porter, K.; Kukkaro, P.; Bamford, J.K.; Bath, C.; Kivela, H.M.; Dyall-Smith, M.L.; Bamford, D.H. Sh1: A novel, spherical halovirus isolated from an australian hypersaline lake. Virology 2005, 335, 22–33. [Google Scholar] [CrossRef]
- Pina, M.; Bize, A.; Forterre, P.; Prangishvili, D. The archeoviruses. FEMS Microbiol. Rev. 2011, 35, 1035–1054. [Google Scholar] [CrossRef]
- Maaty, W.S.; Ortmann, A.C.; Dlakic, M.; Schulstad, K.; Hilmer, J.K.; Liepold, L.; Weidenheft, B.; Khayat, R.; Douglas, T.; Young, M.J.; et al. Characterization of the archaeal thermophile sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J. Virol. 2006, 80, 7625–7635. [Google Scholar] [CrossRef]
- Bettstetter, M.; Peng, X.; Garrett, R.A.; Prangishvili, D. Afv1, a novel virus infecting hyperthermophilic archaea of the genus acidianus. Virology 2003, 315, 68–79. [Google Scholar] [CrossRef]
- Zillig, W.; Kletzin, A.; Schleper, C.; Holz, I.; Janekovic, D.; Hain, J.; Lanzendorfer, M.; Kristjansson, J.K. Screening for sulfolobales, their plasmids and their viruses in icelandic solfataras. Systematic and Appl. Microbiol. 1994, 16, 609–628. [Google Scholar]
- Palm, P.; Schleper, C.; Grampp, B.; Yeats, S.; Mcwilliam, P.; Reiter, W.D.; Zillig, W. Complete nucleotide-sequence of the virus ssv1 of the archaebacterium sulfolobus-shibatae. Virology 1991, 185, 242–250. [Google Scholar] [CrossRef]
- Richardson, J.S. The anatomy and taxonomy of protein structure. Adv. Protein. Chem. 1981, 34, 167–339. [Google Scholar]
- Khayat, R.; Tang, L.; Larson, E.T.; Lawrence, C.M.; Young, M.; Johnson, J.E. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc. Natl. Acad. Sci. USA 2005, 102, 18944–18949. [Google Scholar]
- Benson, S.D.; Bamford, J.K.; Bamford, D.H.; Burnett, R.M. Viral evolution revealed by bacteriophage prd1 and human adenovirus coat protein structures. Cell 1999, 98, 825–833. [Google Scholar] [CrossRef]
- Nandhagopal, N.; Simpson, A.A.; Gurnon, J.R.; Yan, X.; Baker, T.S.; Graves, M.V.; Van Etten, J.L.; Rossmann, M.G. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. USA 2002, 99, 14758–14763. [Google Scholar]
- Stromsten, N.J.; Bamford, D.H.; Bamford, J.K. In vitro DNA packaging of prd1: A common mechanism for internal-membrane viruses. J Mol. Biol. 2005, 348, 617–629. [Google Scholar] [CrossRef]
- Happonen, L.J.; Redder, P.; Peng, X.; Reigstad, L.J.; Prangishvili, D.; Butcher, S.J. Familial relationships in hyperthermo- and acidophilic archaeal viruses. J. Virol. 2010, 84, 4747–4754. [Google Scholar]
- San Martin, C.; Huiskonen, J.T.; Bamford, J.K.; Butcher, S.J.; Fuller, S.D.; Bamford, D.H.; Burnett, R.M. Minor proteins, mobile arms and membrane-capsid interactions in the bacteriophage prd1 capsid. Nat. Struct. Biol. 2002, 9, 756–763. [Google Scholar] [CrossRef]
- Goulet, A.; Blangy, S.; Redder, P.; Prangishvili, D.; Felisberto-Rodrigues, C.; Forterre, P.; Campanacci, V.; Cambillau, C. Acidianus filamentous virus 1 coat proteins display a helical fold spanning the filamentous archaeal viruses lineage. Proc. Natl. Acad. Sci. USA 2009, 106, 21155–21160. [Google Scholar]
- Namba, K.; Stubbs, G. Structure of tobacco mosaic virus at 3.6 a resolution: Implications for assembly. Science 1986, 231, 1401–1406. [Google Scholar]
- Aravind, L.; Anantharaman, V.; Balaji, S.; Babu, M.M.; Iyer, L.M. The many faces of the helix-turn-helix domain: Transcription regulation and beyond. FEMS Microbiol. Rev. 2005, 29, 231–262. [Google Scholar]
- Larson, E.T.; Eilers, B.; Menon, S.; Reiter, D.; Ortmann, A.; Young, M.J.; Lawrence, C.M. A winged-helix protein from sulfolobus turreted icosahedral virus points toward stabilizing disulfide bonds in the intracellular proteins of a hyperthermophilic virus. Virology 2007, 368, 249–261. [Google Scholar]
- Kraft, P.; Oeckinghaus, A.; Kummel, D.; Gauss, G.H.; Gilmore, J.; Wiedenheft, B.; Young, M.; Lawrence, C.M. Crystal structure of f-93 from sulfolobus spindle-shaped virus 1, a winged-helix DNA binding protein. J. Virol. 2004, 78, 11544–11550. [Google Scholar]
- Szymczyna, B.R.; Taurog, R.E.; Young, M.J.; Snyder, J.C.; Johnson, J.E.; Williamson, J.R. Synergy of nmr, computation, and x-ray crystallography for structural biology. Structure 2009, 17, 499–507. [Google Scholar] [CrossRef]
- Schreiter, E.R.; Drennan, C.L. Ribbon-helix-helix transcription factors: Variations on a theme. Nat. Rev. Microbiol. 2007, 5, 710–720. [Google Scholar] [CrossRef]
- Schlenker, C.; Goel, A.; Tripet, B.P.; Menon, S.; Willi, T.; Dlakic, M.; Young, M.J.; Lawrence, C.M.; Copie, V. Structural studies of e73 from a hyperthermophilic archaeal virus identify the "rh3" domain, an elaborated ribbon-helix-helix motif involved in DNA recognition. Biochemistry 2012, 51, 2899–2910. [Google Scholar]
- Guilliere, F.; Peixeiro, N.; Kessler, A.; Raynal, B.; Desnoues, N.; Keller, J.; Delepierre, M.; Prangishvili, D.; Sezonov, G.; Guijarro, J.I. Structure, function, and targets of the transcriptional regulator svtr from the hyperthermophilic archaeal virus sirv1. J. Biol. Chem. 2009, 284, 22222–22237. [Google Scholar]
- Goulet, A.; Pina, M.; Redder, P.; Prangishvili, D.; Vera, L.; Lichiere, J.; Leulliot, N.; van Tilbeurgh, H.; Ortiz-Lombardia, M.; Campanacci, V.; et al. Orf157 from the archaeal virus acidianus filamentous virus 1 defines a new class of nuclease. J. Virol. 2010, 84, 5025–5031. [Google Scholar]
- Menon, S.K.; Eilers, B.J.; Young, M.J.; Lawrence, C.M. The crystal structure of d212 from sulfolobus spindle-shaped virus ragged hills reveals a new member of the pd-(d/e)xk nuclease superfamily. J. Virol. 2010, 84, 5890–5897. [Google Scholar]
- Eilers, B.J.; Young, M.J.; Lawrence, C.M. The structure of an archaeal viral integrase reveals an evolutionarily conserved catalytic core yet supports a mechanism of DNA cleavage in trans. J. Virol. 2012, 86, 8309–8313. [Google Scholar] [CrossRef]
- Zhan, Z.; Ouyang, S.; Liang, W.; Zhang, Z.; Liu, Z.J.; Huang, L. Structural and functional characterization of the c-terminal catalytic domain of ssv1 integrase. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 659–670. [Google Scholar]
- Oke, M.; Kerou, M.; Liu, H.T.; Peng, X.; Garrett, R.A.; Prangishvili, D.; Naismith, J.H.; White, M.F. A dimeric rep protein initiates replication of a linear archaeal virus genome: Implications for the rep mechanism and viral replication. J. Virol. 2011, 85, 925–931. [Google Scholar]
- Larson, E.T.; Reiter, D.; Young, M.; Lawrence, C.M. Structure of a197 from sulfolobus turreted icosahedral virus: A crenarchaeal viral glycosyltransferase exhibiting the gt-a fold. J. Virol. 2006, 80, 7636–7644. [Google Scholar]
- Bond, C.S.; Kvaratskhelia, M.; Richard, D.; White, M.F.; Hunter, W.N. Structure of hjc, a holliday junction resolvase, from sulfolobus solfataricus. Proc. Natl. Acad. Sci. USA 2001, 98, 5509–5514. [Google Scholar]
- Dyda, F.; Hickman, A.B.; Jenkins, T.M.; Engelman, A.; Craigie, R.; Davies, D.R. Crystal structure of the catalytic domain of HIV-1 integrase: Similarity to other polynucleotidyl transferases. Science 1994, 266, 1981–1986. [Google Scholar]
- Letzelter, C.; Duguet, M.; Serre, M.C. Mutational analysis of the archaeal tyrosine recombinase ssv1 integrase suggests a mechanism of DNA cleavage in trans. J. Biol. Chem. 2004, 279, 28936–28944. [Google Scholar] [CrossRef]
- Fu, C.Y.; Wang, K.; Gan, L.; Lanman, J.; Khayat, R.; Young, M.J.; Jensen, G.J.; Doerschuk, P.C.; Johnson, J.E. In vivo assembly of an archaeal virus studied with whole-cell electron cryotomography. Structure 2010, 18, 1579–1586. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. (Eds.) Viruses, Plasmids, and Transposable Genetic Elements. Molecular Biology of the Cell; Garland Science: New York, NY, USA, 1994, 3rd edition Available online: http://www.ncbi.nlm.nih.gov/books/NBK28286/ (accessed on 24 November 2012).
- Goulet, A.; Spinelli, S.; Blangy, S.; van Tilbeurgh, H.; Leulliot, N.; Basta, T.; Prangishvili, D.; Cambillau, C.; Campanacci, V. The thermo- and acido-stable orf-99 from the archaeal virus afv1. Protein Sci. 2009, 18, 1316–1320. [Google Scholar]
- Felisberto-Rodrigues, C.; Blangy, S.; Goulet, A.; Vestergaard, G.; Cambillau, C.; Garrett, R.A.; Ortiz-Lombardia, M. Crystal structure of atv (orf273), a new fold for a thermo- and acido-stable protein from the acidianus two-tailed virus. Plos One 2012. [Google Scholar] [CrossRef]
- Goulet, A.; Spinelli, S.; Blangy, S.; van Tilbeurgh, H.; Leulliot, N.; Basta, T.; Prangishvili, D.; Cambillau, C.; Campanacci, V. The crystal structure of orf14 from sulfolobus islandicus filamentous virus. Proteins 2009, 76, 1020–1022. [Google Scholar] [CrossRef]
- Menon, S.K.; Maaty, W.S.; Corn, G.J.; Kwok, S.C.; Eilers, B.J.; Kraft, P.; Gillitzer, E.; Young, M.J.; Bothner, B.; Lawrence, C.M. Cysteine usage in sulfolobus spindle-shaped virus 1 and extension to hyperthermophilic viruses in general. Virology 2008, 376, 270–278. [Google Scholar]
- Keller, J.; Leulliot, N.; Collinet, B.; Campanacci, V.; Cambillau, C.; Pranghisvilli, D.; van Tilbeurgh, H. Crystal structure of afv1–102, a protein from the acidianus filamentous virus 1. Protein Sci. 2009, 18, 845–849. [Google Scholar]
- Larson, E.T.; Eilers, B.J.; Reiter, D.; Ortmann, A.C.; Young, M.J.; Lawrence, C.M. A new DNA binding protein highly conserved in diverse crenarchaeal viruses. Virology 2007, 363, 387–396. [Google Scholar] [CrossRef]
- Beeby, M.; O'Connor, B.D.; Ryttersgaard, C.; Boutz, D.R.; Perry, L.J.; Yeates, T.O. The genomics of disulfide bonding and protein stabilization in thermophiles. PLoS Biol 2005, 3. [Google Scholar] [CrossRef] [Green Version]
- Mallick, P.; Boutz, D.R.; Eisenberg, D.; Yeates, T.O. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc. Natl Acad Sci USA 2002, 99, 9679–9684. [Google Scholar]
- Jorda, J.; Yeates, T.O. Widespread disulfide bonding in proteins from thermophilic archaea. Archaea 2011, 2011, 409156. [Google Scholar]
- Keller, J.; Leulliot, N.; Cambillau, C.; Campanacci, V.; Porciero, S.; Prangishvili, D.; Forterre, P.; Cortez, D.; Quevillon-Cheruel, S.; van Tilbeurgh, H. Crystal structure of afv3–109, a highly conserved protein from crenarchaeal viruses. Virol. J. 2007, 4. [Google Scholar] [CrossRef]
- Oke, M.; Carter, L.G.; Johnson, K.A.; Liu, H.; McMahon, S.A.; Yan, X.; Kerou, M.; Weikart, N.D.; Kadi, N.; Sheikh, M.A.; et al. The scottish structural proteomics facility: Targets, methods and outputs. J. Struct. Funct. Genomics 2010, 11, 167–180. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Dellas, N.; Lawrence, C.M.; Young, M.J. A Survey of Protein Structures from Archaeal Viruses. Life 2013, 3, 118-130. https://doi.org/10.3390/life3010118
Dellas N, Lawrence CM, Young MJ. A Survey of Protein Structures from Archaeal Viruses. Life. 2013; 3(1):118-130. https://doi.org/10.3390/life3010118
Chicago/Turabian StyleDellas, Nikki, C. Martin Lawrence, and Mark J. Young. 2013. "A Survey of Protein Structures from Archaeal Viruses" Life 3, no. 1: 118-130. https://doi.org/10.3390/life3010118
APA StyleDellas, N., Lawrence, C. M., & Young, M. J. (2013). A Survey of Protein Structures from Archaeal Viruses. Life, 3(1), 118-130. https://doi.org/10.3390/life3010118





