Lifestyle-Driven Variations in Nutrimiromic MicroRNA Expression Patterns across and beyond Genders
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Group
2.2. Questionnaire
2.3. Sample Collection
2.4. cDNA-Synthesis and Real-Time PCR (qPCR)
2.5. Statistical Analysis
3. Results
3.1. Age-Related Variations in MiRNA Expression
3.2. BMI Effect on MiRNA Levels
3.3. MiRNA Expression across Different Health Statuses
3.4. Impact of Smoking Behaviour on MiRNA Expression
3.5. Alcohol Consumption and Effects on MiRNA Expression
3.6. MiRNA Expression across Different Diet Clusters
3.7. Impact of Milk Consumption on MiRNA Profiles
3.8. Effects of Meat Consumption on MiRNA Expression
3.9. Impact of Vegetable and Fruit Intake on MiRNA Expression
3.10. Sex-Biased MiRNA Expression
4. Discussion
4.1. MiRNAs in Metabolic Diseases and Obesity
4.2. MiRNAs and Ageing
4.3. Impact of Lifestyle Choices: Smoking, Alcohol, and miRNA Expression
4.4. Diet and Nutrimiromics
4.5. Gender-Specific Differences, Immune System, and Personalized Interventions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Asghar, W.; Khalid, N. Nutrigenetics and Nutrigenomics, and Precision Nutrition. Nutr. Health 2023, 29, 169–170. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Martinez, J.A.; Milagro, F.I. Holistic Integration of Omics Tools for Precision Nutrition in Health and Disease. Nutrients 2022, 14, 4074. [Google Scholar] [CrossRef] [PubMed]
- Missong, H.; Joshi, R.; Khullar, N.; Thareja, S.; Navik, U.; Bhatti, G.K.; Bhatti, J.S. Nutrient-Epigenome Interactions: Implications for Personalized Nutrition against Aging-Associated Diseases. J. Nutr. Biochem. 2024, 127, 109592. [Google Scholar] [CrossRef] [PubMed]
- Berciano, S.; Figueiredo, J.; Brisbois, T.D.; Alford, S.; Koecher, K.; Eckhouse, S.; Ciati, R.; Kussmann, M.; Ordovas, J.M.; Stebbins, K.; et al. Precision Nutrition: Maintaining Scientific Integrity While Realizing Market Potential. Front. Nutr. 2022, 9, 979665. [Google Scholar] [CrossRef]
- Manuel, R.S.J.; Liang, Y. Sexual Dimorphism in Immunometabolism and Autoimmunity: Impact on Personalized Medicine. Autoimmun. Rev. 2021, 20, 102775. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, C.; Koivumäki, J.; Pekkanen-Mattila, M.; Aalto-Setälä, K. Sex Differences in Heart: From Basics to Clinics. Eur. J. Med. Res. 2022, 27, 241. [Google Scholar] [CrossRef] [PubMed]
- Lipoldová, M.; Demant, P. Gene-Specific Sex Effects on Susceptibility to Infectious Diseases. Front. Immunol. 2021, 12, 712688. [Google Scholar] [CrossRef]
- Brennan, L.; de Roos, B. Nutrigenomics: Lessons Learned and Future Perspectives. Am. J. Clin. Nutr. 2021, 113, 503–516. [Google Scholar] [CrossRef]
- Abdellaoui, A.; Yengo, L.; Verweij, K.J.H.; Visscher, P.M. 15 Years of GWAS Discovery: Realizing the Promise. Am. J. Hum. Genet. 2023, 110, 179–194. [Google Scholar] [CrossRef]
- Sud, A.; Kinnersley, B.; Houlston, R.S. Genome-Wide Association Studies of Cancer: Current Insights and Future Perspectives. Nat. Rev. Cancer 2017, 17, 692–704. [Google Scholar] [CrossRef]
- Oo, J.A.; Brandes, R.P.; Leisegang, M.S. Long Non-Coding RNAs: Novel Regulators of Cellular Physiology and Function. Pflugers Arch. 2022, 474, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Quinodoz, S.; Guttman, M. Long Noncoding RNAs: An Emerging Link between Gene Regulation and Nuclear Organization. Trends Cell Biol. 2014, 24, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC Couples MicroRNA Biogenesis and Posttranscriptional Gene Silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Elshazli, R.M.; Toraih, E.A.; Hussein, M.H.; Ruiz, E.M.; Kandil, E.; Fawzy, M.S. Pan-Cancer Study on Variants of Canonical miRNA Biogenesis Pathway Components: A Pooled Analysis. Cancers 2023, 15, 338. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of Functional microRNA Targets by Integrative Modeling of microRNA Binding and Target Expression Data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Raisch, J.; Darfeuille-Michaud, A.; Nguyen, H.T.T. Role of microRNAs in the Immune System, Inflammation and Cancer. World J. Gastroenterol. 2013, 19, 2985–2996. [Google Scholar] [CrossRef]
- Zhou, H.; Ni, W.-J.; Meng, X.-M.; Tang, L.-Q. MicroRNAs as Regulators of Immune and Inflammatory Responses: Potential Therapeutic Targets in Diabetic Nephropathy. Front. Cell Dev. Biol. 2021, 8, 618536. [Google Scholar] [CrossRef] [PubMed]
- Oses, M.; Margareto Sanchez, J.; Portillo, M.P.; Aguilera, C.M.; Labayen, I. Circulating miRNAs as Biomarkers of Obesity and Obesity-Associated Comorbidities in Children and Adolescents: A Systematic Review. Nutrients 2019, 11, 2890. [Google Scholar] [CrossRef]
- Roy, B.; Lee, E.; Li, T.; Rampersaud, M. Role of miRNAs in Neurodegeneration: From Disease Cause to Tools of Biomarker Discovery and Therapeutics. Genes 2022, 13, 425. [Google Scholar] [CrossRef]
- Mastropasqua, R.; D’Aloisio, R.; Costantini, E.; Porreca, A.; Ferro, G.; Libertini, D.; Reale, M.; Di Nicola, M.; Viggiano, P.; Falconio, G.; et al. Serum microRNA Levels in Diabetes Mellitus. Diagnostics 2021, 11, 284. [Google Scholar] [CrossRef]
- Li, X.; Dai, A.; Tran, R.; Wang, J. Identifying miRNA Biomarkers for Breast Cancer and Ovarian Cancer: A Text Mining Perspective. Breast Cancer Res. Treat. 2023, 201, 5–14. [Google Scholar] [CrossRef]
- Xu, W.-D.; Feng, S.-Y.; Huang, A.-F. Role of miR-155 in Inflammatory Autoimmune Diseases: A Comprehensive Review. Inflamm. Res. 2022, 71, 1501–1517. [Google Scholar] [CrossRef] [PubMed]
- Krammer, U.D.B.; Tschida, S.; Berner, J.; Lilja, S.; Switzeny, O.J.; Hippe, B.; Rust, P.; Haslberger, A.G. MiRNA-Based “Fitness Score” to Assess the Individual Response to Diet, Metabolism, and Exercise. J. Int. Soc. Sports Nutr. 2022, 19, 455–473. [Google Scholar] [CrossRef] [PubMed]
- Vrijens, K.; Bollati, V.; Nawrot, T.S. MicroRNAs as Potential Signatures of Environmental Exposure or Effect: A Systematic Review. Environ. Health Perspect. 2015, 123, 399–411. [Google Scholar] [CrossRef]
- Zhou, Q.; Shi, C.; Lv, Y.; Zhao, C.; Jiao, Z.; Wang, T. Circulating microRNAs in Response to Exercise Training in Healthy Adults. Front. Genet. 2020, 11, 256. [Google Scholar] [CrossRef] [PubMed]
- Willinger, C.M.; Rong, J.; Tanriverdi, K.; Courchesne, P.L.; Huan, T.; Wasserman, G.A.; Lin, H.; Dupuis, J.; Joehanes, R.; Jones, M.R.; et al. A MicroRNA Signature of Cigarette Smoking and Evidence for a Putative Causal Role of MicroRNAs in Smoking-Related Inflammation and Target Organ Damage. Circ. Cardiovasc. Genet. 2017, 10, e001678. [Google Scholar] [CrossRef] [PubMed]
- Kura, B.; Parikh, M.; Slezak, J.; Pierce, G.N. The Influence of Diet on MicroRNAs That Impact Cardiovascular Disease. Molecules 2019, 24, 1509. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as Potential Cancer Biomarkers: The Advantage and Disadvantage. Clin. Epigenet. 2018, 10, 59. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as Stable Blood-Based Markers for Cancer Detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Chirshev, E.; Oberg, K.C.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7 as Biomarker, Prognostic Indicator, and Therapy for Precision Medicine in Cancer. Clin. Transl. Med. 2019, 8, 24. [Google Scholar] [CrossRef]
- Biamonte, F.; Santamaria, G.; Sacco, A.; Perrone, F.M.; Di Cello, A.; Battaglia, A.M.; Salatino, A.; Di Vito, A.; Aversa, I.; Venturella, R.; et al. MicroRNA Let-7g Acts as Tumor Suppressor and Predictive Biomarker for Chemoresistance in Human Epithelial Ovarian Cancer. Sci. Rep. 2019, 9, 5668. [Google Scholar] [CrossRef] [PubMed]
- Aqeilan, R.I.; Calin, G.A.; Croce, C.M. miR-15a and miR-16-1 in Cancer: Discovery, Function and Future Perspectives. Cell Death Differ. 2010, 17, 215–220. [Google Scholar] [CrossRef]
- Hong, Y.; He, H.; Jiang, G.; Zhang, H.; Tao, W.; Ding, Y.; Yuan, D.; Liu, J.; Fan, H.; Lin, F.; et al. miR-155-5p Inhibition Rejuvenates Aged Mesenchymal Stem Cells and Enhances Cardioprotection Following Infarction. Aging Cell 2020, 19, e13128. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Prattichizzo, F.; Giuliani, A.; Matacchione, G.; Rippo, M.R.; Sabbatinelli, J.; Bonafè, M. miR-21 and miR-146a: The microRNAs of Inflammaging and Age-Related Diseases. Ageing Res. Rev. 2021, 70, 101374. [Google Scholar] [CrossRef]
- Huang, X.-Y.; Chen, J.-X.; Ren, Y.; Fan, L.-C.; Xiang, W.; He, X.-J. Exosomal miR-122 Promotes Adipogenesis and Aggravates Obesity through the VDR/SREBF1 Axis. Obesity 2022, 30, 666–679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, S.; Jia, Q.; Zhang, A.; Li, Y.; Zhu, Y.; Lv, S.; Zhang, J. The microRNA in Ventricular Remodeling: The miR-30 Family. Biosci. Rep. 2019, 39, BSR20190788. [Google Scholar] [CrossRef] [PubMed]
- Thermo Fisher Scientific. A Technical Guide to Identifying miRNA Normalizers Using TaqMan Advanced miRNA Assays; Thermo Fisher Scientific: Waltham, MA, USA, 2016. [Google Scholar]
- Iacomino, G.; Siani, A. Role of microRNAs in Obesity and Obesity-Related Diseases. Genes Nutr. 2017, 12, 23. [Google Scholar] [CrossRef]
- Loganathan, T.; Doss, C.G.P. Non-Coding RNAs in Human Health and Disease: Potential Function as Biomarkers and Therapeutic Targets. Funct. Integr. Genom. 2023, 23, 33. [Google Scholar] [CrossRef]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-Based Biomarkers, Therapies, and Resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef]
- Cui, C.; Cui, Q. The Relationship of Human Tissue microRNAs with Those from Body Fluids. Sci. Rep. 2020, 10, 5644. [Google Scholar] [CrossRef]
- Waters, P.S.; McDermott, A.M.; Wall, D.; Heneghan, H.M.; Miller, N.; Newell, J.; Kerin, M.J.; Dwyer, R.M. Relationship between Circulating and Tissue microRNAs in a Murine Model of Breast Cancer. PLoS ONE 2012, 7, e50459. [Google Scholar] [CrossRef] [PubMed]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-Derived Circulating miRNAs Regulate Gene Expression in Other Tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Rakib, A.; Kiran, S.; Mandal, M.; Singh, U.P. MicroRNAs: A Crossroad That Connects Obesity to Immunity and Aging. Immun. Ageing 2022, 19, 64. [Google Scholar] [CrossRef] [PubMed]
- Silveira, A.; Gomes, J.; Roque, F.; Fernandes, T.; de Oliveira, E.M. MicroRNAs in Obesity-Associated Disorders: The Role of Exercise Training. Obes. Facts 2022, 15, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Deiuliis, J.A. MicroRNAs as Regulators of Metabolic Disease: Pathophysiologic Significance and Emerging Role as Biomarkers and Therapeutics. Int. J. Obes. 2016, 40, 88–101. [Google Scholar] [CrossRef] [PubMed]
- Bulló, M.; García-Lorda, P.; Megias, I.; Salas-Salvadó, J. Systemic Inflammation, Adipose Tissue Tumor Necrosis Factor, and Leptin Expression. Obes. Res. 2003, 11, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Mabrouk, R.; Ghareeb, H.; Shehab, A.; Omar, K.; El-Kabarity, R.H.; Soliman, D.A.; Mohamed, N.A. Serum Visfatin, Resistin and IL-18 in A Group of Egyptian Obese Diabetic and Non Diabetic Individuals. Egypt. J. Immunol. 2013, 20, 1–11. [Google Scholar]
- Festa, A.; D’Agostino, R.; Williams, K.; Karter, A.J.; Mayer-Davis, E.J.; Tracy, R.P.; Haffner, S.M. The Relation of Body Fat Mass and Distribution to Markers of Chronic Inflammation. Int. J. Obes. Relat. Metab. Disord. J. Int. Assoc. Study Obes. 2001, 25, 1407–1415. [Google Scholar] [CrossRef]
- Clemente-Suárez, V.J.; Redondo-Flórez, L.; Beltrán-Velasco, A.I.; Martín-Rodríguez, A.; Martínez-Guardado, I.; Navarro-Jiménez, E.; Laborde-Cárdenas, C.C.; Tornero-Aguilera, J.F. The Role of Adipokines in Health and Disease. Biomedicines 2023, 11, 1290. [Google Scholar] [CrossRef]
- Kim, Y.J.; Hwang, S.J.; Bae, Y.C.; Jung, J.S. MiR-21 Regulates Adipogenic Differentiation through the Modulation of TGF-Beta Signaling in Mesenchymal Stem Cells Derived from Human Adipose Tissue. Stem Cells 2009, 27, 3093–3102. [Google Scholar] [CrossRef]
- Keller, P.; Gburcik, V.; Petrovic, N.; Gallagher, I.J.; Nedergaard, J.; Cannon, B.; Timmons, J.A. Gene-Chip Studies of Adipogenesis-Regulated microRNAs in Mouse Primary Adipocytes and Human Obesity. BMC Endocr. Disord. 2011, 11, 7. [Google Scholar] [CrossRef]
- Lhamyani, S.; Gentile, A.-M.; Giráldez-Pérez, R.M.; Feijóo-Cuaresma, M.; Romero-Zerbo, S.Y.; Clemente-Postigo, M.; Zayed, H.; Oliva-Olivera, W.; Bermúdez-Silva, F.J.; Salas, J.; et al. miR-21 Mimic Blocks Obesity in Mice: A Novel Therapeutic Option. Mol. Ther.—Nucleic Acids 2021, 26, 401–416. [Google Scholar] [CrossRef] [PubMed]
- Yazarlou, F.; Kadkhoda, S.; Ghafouri-Fard, S. Emerging Role of Let-7 Family in the Pathogenesis of Hematological Malignancies. Biomed. Pharmacother. 2021, 144, 112334. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Huang, Y.; Zhao, J.-X.; Rogers, C.J.; Zhu, M.-J.; Ford, S.P.; Nathanielsz, P.W.; Du, M. Maternal Obesity Downregulates microRNA Let-7g Expression, a Possible Mechanism for Enhanced Adipogenesis during Ovine Fetal Skeletal Muscle Development. Int. J. Obes. 2013, 37, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Karkeni, E.; Astier, J.; Tourniaire, F.; El Abed, M.; Romier, B.; Gouranton, E.; Wan, L.; Borel, P.; Salles, J.; Walrand, S.; et al. Obesity-Associated Inflammation Induces microRNA-155 Expression in Adipocytes and Adipose Tissue: Outcome on Adipocyte Function. J. Clin. Endocrinol. Metab. 2016, 101, 1615–1626. [Google Scholar] [CrossRef] [PubMed]
- Jankauskas, S.S.; Gambardella, J.; Sardu, C.; Lombardi, A.; Santulli, G. Functional Role of miR-155 in the Pathogenesis of Diabetes Mellitus and Its Complications. Non-Coding RNA 2021, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Drummer, C.; Virtue, A.; Gao, T.; Wu, S.; Hernandez, M.; Singh, L.; Wang, H.; Yang, X.-F. Increased Expression of Resistin in MicroRNA-155-Deficient White Adipose Tissues May Be a Possible Driver of Metabolically Healthy Obesity Transition to Classical Obesity. Front. Physiol. 2018, 9, 1297. [Google Scholar] [CrossRef]
- Corral-Fernández, N.E.; Salgado-Bustamante, M.; Martínez-Leija, M.E.; Cortez-Espinosa, N.; García-Hernández, M.H.; Reynaga-Hernández, E.; Quezada-Calvillo, R.; Portales-Pérez, D.P. Dysregulated miR-155 Expression in Peripheral Blood Mononuclear Cells from Patients with Type 2 Diabetes. Exp. Clin. Endocrinol. Diabetes 2013, 121, 347–353. [Google Scholar] [CrossRef]
- Mazloom, H.; Alizadeh, S.; Pasalar, P.; Esfahani, E.N.; Meshkani, R. Downregulated microRNA-155 Expression in Peripheral Blood Mononuclear Cells of Type 2 Diabetic Patients Is Not Correlated with Increased Inflammatory Cytokine Production. Cytokine 2015, 76, 403–408. [Google Scholar] [CrossRef]
- Ciccacci, C.; Latini, A.; Colantuono, A.; Politi, C.; D’Amato, C.; Greco, C.; Rinaldi, M.E.; Lauro, D.; Novelli, G.; Spallone, V.; et al. Expression Study of Candidate miRNAs and Evaluation of Their Potential Use as Biomarkers of Diabetic Neuropathy. Epigenomics 2020, 12, 575–585. [Google Scholar] [CrossRef]
- Tryggestad, J.B.; Teague, A.M.; Sparling, D.P.; Jiang, S.; Chernausek, S.D. Macrophage Derived MicroRNA-155 Increases in Obesity and Influences Adipocyte Metabolism by Targeting Peroxisome Proliferator-Activated Receptor Gamma. Obesity 2019, 27, 1856–1864. [Google Scholar] [CrossRef]
- Catanzaro, G.; Conte, F.; Trocchianesi, S.; Splendiani, E.; Bimonte, V.M.; Mocini, E.; Filardi, T.; Po, A.; Besharat, Z.M.; Gentile, M.C.; et al. Network Analysis Identifies Circulating miR-155 as Predictive Biomarker of Type 2 Diabetes Mellitus Development in Obese Patients: A Pilot Study. Sci. Rep. 2023, 13, 19496. [Google Scholar] [CrossRef] [PubMed]
- Merve Bayram, H.; Eren, F.; Esra Gunes, F. The Relationship between Polyphenols and miRNAs: A Novel Therapeutic Strategy for Metabolic Associated Fatty Liver Disease. Hepatol. Forum 2021, 2, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Kanigur Sultuybek, G.; Soydas, T.; Yenmis, G. NF-κB as the Mediator of Metformin’s Effect on Ageing and Ageing-Related Diseases. Clin. Exp. Pharmacol. Physiol. 2019, 46, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Al-Kafaji, G.; Al-Mahroos, G.; Alsayed, N.A.; Hasan, Z.A.; Nawaz, S.; Bakhiet, M. Peripheral Blood microRNA-15a Is a Potential Biomarker for Type 2 Diabetes Mellitus and Pre-Diabetes. Mol. Med. Rep. 2015, 12, 7485–7490. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.E.; Brown, J.L.; Rosa, M.E.; Brown, L.A.; Perry, R.A.; Wiggs, M.P.; Nilsson, M.I.; Crouse, S.F.; Fluckey, J.D.; Washington, T.A.; et al. microRNA-16 Is Downregulated During Insulin Resistance and Controls Skeletal Muscle Protein Accretion. J. Cell. Biochem. 2016, 117, 1775–1787. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, L.; Fabbri, E. Inflammageing: Chronic Inflammation in Ageing, Cardiovascular Disease, and Frailty. Nat. Rev. Cardiol. 2018, 15, 505–522. [Google Scholar] [CrossRef] [PubMed]
- Montecino-Rodriguez, E.; Berent-Maoz, B.; Dorshkind, K. Causes, Consequences, and Reversal of Immune System Aging. J. Clin. Investig. 2013, 123, 958–965. [Google Scholar] [CrossRef]
- Harries, L.W. MicroRNAs as Mediators of the Ageing Process. Genes 2014, 5, 656–670. [Google Scholar] [CrossRef]
- Borja-Gonzalez, M.; Casas-Martinez, J.C.; McDonagh, B.; Goljanek-Whysall, K. Inflamma-miR-21 Negatively Regulates Myogenesis during Ageing. Antioxidants 2020, 9, 345. [Google Scholar] [CrossRef]
- Guedes, J.R.; Custódia, C.M.; Silva, R.J.; de Almeida, L.P.; Pedroso de Lima, M.C.; Cardoso, A.L. Early miR-155 Upregulation Contributes to Neuroinflammation in Alzheimer’s Disease Triple Transgenic Mouse Model. Hum. Mol. Genet. 2014, 23, 6286–6301. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Shen, N.; Wicha, M.S.; Luo, M. The Roles of the Let-7 Family of MicroRNAs in the Regulation of Cancer Stemness. Cells 2021, 10, 2415. [Google Scholar] [CrossRef] [PubMed]
- Wells, A.C.; Hioki, K.A.; Angelou, C.C.; Lynch, A.C.; Liang, X.; Ryan, D.J.; Thesmar, I.; Zhanybekova, S.; Zuklys, S.; Ullom, J.; et al. Let-7 Enhances Murine Anti-Tumor CD8 T Cell Responses by Promoting Memory and Antagonizing Terminal Differentiation. Nat. Commun. 2023, 14, 5585. [Google Scholar] [CrossRef]
- Tristán-Ramos, P.; Rubio-Roldan, A.; Peris, G.; Sánchez, L.; Amador-Cubero, S.; Viollet, S.; Cristofari, G.; Heras, S.R. The Tumor Suppressor microRNA Let-7 Inhibits Human LINE-1 Retrotransposition. Nat. Commun. 2020, 11, 5712. [Google Scholar] [CrossRef] [PubMed]
- Pulliero, A.; Mastracci, L.; Tarantini, L.; Khalid, Z.; Bollati, V.; Izzotti, A. Let-7a Downregulation Accompanied by KRAS Mutation Is Predictive of Lung Cancer Onset in Cigarette Smoke-Exposed Mice. Int. J. Mol. Sci. 2023, 24, 11778. [Google Scholar] [CrossRef] [PubMed]
- Asghari, F.; Haghnavaz, N.; Shanehbandi, D.; Khaze, V.; Baradaran, B.; Kazemi, T. Differential Altered Expression of Let-7a and miR-205 Tumor-Suppressor miRNAs in Different Subtypes of Breast Cancer under Treatment with Taxol. Adv. Clin. Exp. Med. 2018, 27, 941–945. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.J.; Song, J.; Oh, Y.; Lee, J.E. MicroRNA-Let-7a Regulates the Function of Microglia in Inflammation. Mol. Cell. Neurosci. 2015, 68, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yokota, S.; Tatsumi, N.; Fukami, T.; Yokoi, T.; Nakajima, M. Cigarette Smoking Substantially Alters Plasma microRNA Profiles in Healthy Subjects. Toxicol. Appl. Pharmacol. 2013, 272, 154–160. [Google Scholar] [CrossRef]
- Karabegović, I.; Maas, S.C.E.; Shuai, Y.; Ikram, M.A.; Stricker, B.; Aerts, J.; Brusselle, G.; Lahousse, L.; Voortman, T.; Ghanbari, M. Smoking-Related Dysregulation of Plasma Circulating microRNAs: The Rotterdam Study. Hum. Genom. 2023, 17, 61. [Google Scholar] [CrossRef]
- Wu, X.-B.; Li, Q.-H.; Zhang, N.; Li, M.; Li, K. MiR-142 Inhibits Lung Cancer Cell Proliferation and Promotes Apoptosis by Targeting XIAP. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7430–7437. [Google Scholar] [CrossRef]
- Yan, J.; Yang, B.; Lin, S.; Xing, R.; Lu, Y. Downregulation of miR-142-5p Promotes Tumor Metastasis through Directly Regulating CYR61 Expression in Gastric Cancer. Gastric Cancer Off. J. Int. Gastric Cancer Assoc. Jpn. Gastric Cancer Assoc. 2019, 22, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; He, F.; Zhang, M.; Ai, L.; He, M.; Liu, X.; Li, Y. MiR-142-3p as an Indicator of OSA Severity Predicts Prognosis in Lung Adenocarcinoma with OSA. Nat. Sci. Sleep 2022, 14, 2047–2054. [Google Scholar] [CrossRef]
- Aonuma, T.; Moukette, B.; Kawaguchi, S.; Barupala, N.P.; Sepúlveda, M.N.; Corr, C.; Tang, Y.; Liangpunsakul, S.; Payne, R.M.; Willis, M.S.; et al. Cardiomyocyte microRNA-150 Confers Cardiac Protection and Directly Represses Proapoptotic Small Proline-Rich Protein 1A. JCI Insight 2021, 6, e150405. [Google Scholar] [CrossRef] [PubMed]
- Stavast, C.J.; van Zuijen, I.; Erkeland, S.J. MicroRNA-139, an Emerging Gate-Keeper in Various Types of Cancer. Cells 2022, 11, 769. [Google Scholar] [CrossRef] [PubMed]
- Quintanilha, B.J.; Reis, B.Z.; Duarte, G.B.S.; Cozzolino, S.M.F.; Rogero, M.M. Nutrimiromics: Role of microRNAs and Nutrition in Modulating Inflammation and Chronic Diseases. Nutrients 2017, 9, 1168. [Google Scholar] [CrossRef]
- Li, H.; Jia, Z.; Li, A.; Jenkins, G.; Yang, X.; Hu, J.; Guo, W. Resveratrol Repressed Viability of U251 Cells by miR-21 Inhibiting of NF-κB Pathway. Mol. Cell. Biochem. 2013, 382, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Moon, R.; Thorne, J.L.; Moore, J.B. NAFLD and Vitamin D: Evidence for Intersection of microRNA-Regulated Pathways. Nutr. Res. Rev. 2023, 36, 120–139. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Dong, Y.-L.; Li, T.; Xiong, W.; Zhang, X.; Wang, P.-J.; Huang, J.-Q. Dietary Selenium Regulates microRNAs in Metabolic Disease: Recent Progress. Nutrients 2021, 13, 1527. [Google Scholar] [CrossRef]
- Ibrahim, S.S.A.; Kandil, L.S.; Ragab, G.M.; El-Sayyad, S.M. Micro RNAs 26b, 20a Inversely Correlate with GSK-3 β/NF-κB/NLRP-3 Pathway to Highlight the Additive Promising Effects of Atorvastatin and Quercetin in Experimental Induced Arthritis. Int. Immunopharmacol. 2021, 99, 108042. [Google Scholar] [CrossRef]
- Majidinia, M.; Karimian, A.; Alemi, F.; Yousefi, B.; Safa, A. Targeting miRNAs by Polyphenols: Novel Therapeutic Strategy for Aging. Biochem. Pharmacol. 2020, 173, 113688. [Google Scholar] [CrossRef]
- Roessler, C.; Kuhlmann, K.; Hellwing, C.; Leimert, A.; Schumann, J. Impact of Polyunsaturated Fatty Acids on miRNA Profiles of Monocytes/Macrophages and Endothelial Cells—A Pilot Study. Int. J. Mol. Sci. 2017, 18, 284. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Sasano, T.; Sugiyama, K.; Kurokawa, J.; Tamura, N.; Soejima, Y.; Sawabe, M.; Isobe, M.; Furukawa, T. High-Fat Diet Increases Vulnerability to Atrial Arrhythmia by Conduction Disturbance via miR-27b. J. Mol. Cell. Cardiol. 2016, 90, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Iwańczyk, S.; Lehmann, T.; Cieślewicz, A.; Malesza, K.; Woźniak, P.; Hertel, A.; Krupka, G.; Jagodziński, P.P.; Grygier, M.; Lesiak, M.; et al. Circulating miRNA-451a and miRNA-328-3p as Potential Markers of Coronary Artery Aneurysmal Disease. Int. J. Mol. Sci. 2023, 24, 5817. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Z.; Li, J.; Liu, B.-Z. MicroRNA-328-3p Inhibits Malignant Progression of Hepatocellular Carcinoma by Regulating MMP-9 Level. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9331–9340. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Ma, C.; Zhou, N.; Li, X.; Zhang, Y. Up- Regulation of miR-328-3p Sensitizes Non-Small Cell Lung Cancer to Radiotherapy. Sci. Rep. 2016, 6, 31651. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Banerjee, A.; Cui, T.; Han, C.; Cai, S.; Liu, L.; Wu, D.; Cui, R.; Li, Z.; Zhang, X.; et al. Inhibition of miR-328–3p Impairs Cancer Stem Cell Function and Prevents Metastasis in Ovarian Cancer. Cancer Res. 2019, 79, 2314–2326. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Ye, X.-X. MicroRNA-328-3p Inhibits the Tumorigenesis of Bladder Cancer through Targeting ITGA5 and Inactivating PI3K/AKT Pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5139–5148. [Google Scholar] [CrossRef]
- Baier, S.R.; Nguyen, C.; Xie, F.; Wood, J.R.; Zempleni, J. MicroRNAs Are Absorbed in Biologically Meaningful Amounts from Nutritionally Relevant Doses of Cow Milk and Affect Gene Expression in Peripheral Blood Mononuclear Cells, HEK-293 Kidney Cell Cultures, and Mouse Livers123. J. Nutr. 2014, 144, 1495–1500. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous Plant MIR168a Specifically Targets Mammalian LDLRAP1: Evidence of Cross-Kingdom Regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef]
- Van Herwijnen, M.J.C.; Driedonks, T.A.P.; Snoek, B.L.; Kroon, A.M.T.; Kleinjan, M.; Jorritsma, R.; Pieterse, C.M.J.; Hoen, E.N.M.N.-‘t.; Wauben, M.H.M. Abundantly Present miRNAs in Milk-Derived Extracellular Vesicles Are Conserved Between Mammals. Front. Nutr. 2018, 5, 81. [Google Scholar] [CrossRef]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine Milk Contains microRNA and Messenger RNA That Are Stable under Degradative Conditions. J. Dairy Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef] [PubMed]
- Pieters, B.C.H.; Arntz, O.J.; Bennink, M.B.; Broeren, M.G.A.; van Caam, A.P.M.; Koenders, M.I.; van Lent, P.L.E.M.; van den Berg, W.B.; de Vries, M.; van der Kraan, P.M.; et al. Commercial Cow Milk Contains Physically Stable Extracellular Vesicles Expressing Immunoregulatory TGF-β. PLoS ONE 2015, 10, e0121123. [Google Scholar] [CrossRef] [PubMed]
- Abou el qassim, L.; Martínez, B.; Rodríguez, A.; Dávalos, A.; López de las Hazas, M.-C.; Menéndez Miranda, M.; Royo, L.J. Effects of Cow’s Milk Processing on MicroRNA Levels. Foods 2023, 12, 2950. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Miao, Y.; Shan, Y.; Liu, B.; Li, Y.; Zhao, L.; Jia, L. MiR-106b and miR-93 Regulate Cell Progression by Suppression of PTEN via PI3K/Akt Pathway in Breast Cancer. Cell Death Dis. 2017, 8, e2796. [Google Scholar] [CrossRef] [PubMed]
- Khorraminezhad, L.; Rudkowska, I. Dairy Product Intake Modifies MicroRNA Expression among Individuals with Hyperinsulinemia: A Post-Intervention Cross-Sectional Study. Lifestyle Genom. 2022, 15, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Hlavna, M.; Raudenska, M.; Hudcova, K.; Gumulec, J.; Sztalmachova, M.; Tanhäuserova, V.; Babula, P.; Adam, V.; Eckschlager, T.; Kizek, R.; et al. MicroRNAs and Zinc Metabolism-Related Gene Expression in Prostate Cancer Cell Lines Treated with Zinc(II) Ions. Int. J. Oncol. 2012, 41, 2237–2244. [Google Scholar] [CrossRef] [PubMed]
- Shkembi, B.; Huppertz, T. Influence of Dairy Products on Bioavailability of Zinc from Other Food Products: A Review of Complementarity at a Meal Level. Nutrients 2021, 13, 4253. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Ali, A.; Hadlich, F.; Oster, M.; Reyer, H.; Trakooljul, N.; Sommerfeld, V.; Rodehutscord, M.; Wimmers, K.; Ponsuksili, S. Dietary Phosphorus and Calcium in Feed Affects miRNA Profiles and Their mRNA Targets in Jejunum of Two Strains of Laying Hens. Sci. Rep. 2021, 11, 13534. [Google Scholar] [CrossRef]
- Su, Q.; Liu, Y.; Lv, X.-W.; Dai, R.-X.; Yang, X.-H.; Kong, B.-H. LncRNA TUG1 Mediates Ischemic Myocardial Injury by Targeting miR-132-3p/HDAC3 Axis. Am. J. Physiol. Heart Circ. Physiol. 2020, 318, H332–H344. [Google Scholar] [CrossRef]
- Gong, X.; Huang, M.; Chen, L. Mechanism of miR-132-3p Promoting Neuroinflammation and Dopaminergic Neurodegeneration in Parkinson’s Disease. eNeuro 2022, 9, ENEURO.0393-21.2021. [Google Scholar] [CrossRef]
- DiMarco, M.; Zhao, H.; Boulicault, M.; Richardson, S.S. Why “Sex as a Biological Variable” Conflicts with Precision Medicine Initiatives. Cell Rep. Med. 2022, 3, 100550. [Google Scholar] [CrossRef] [PubMed]
- Legato, M.J.; Johnson, P.A.; Manson, J.E. Consideration of Sex Differences in Medicine to Improve Health Care and Patient Outcomes. JAMA 2016, 316, 1865–1866. [Google Scholar] [CrossRef] [PubMed]
- Miller, V.M.; Rocca, W.A.; Faubion, S.S. Sex Differences Research, Precision Medicine, and the Future of Women’s Health. J. Women’s Health 2015, 24, 969–971. [Google Scholar] [CrossRef] [PubMed]
- Christou, E.A.A.; Banos, A.; Kosmara, D.; Bertsias, G.; Boumpas, D. Sexual Dimorphism in SLE: Above and beyond Sex Hormones. Lupus 2019, 28, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Deny, M.; Popotas, A.; Hanssens, L.; Lefèvre, N.; Arroba Nuñez, L.A.; Ouafo, G.S.; Corazza, F.; Casimir, G.; Chamekh, M. Sex-Biased Expression of Selected Chromosome x-Linked microRNAs with Potent Regulatory Effect on the Inflammatory Response in Children with Cystic Fibrosis: A Preliminary Pilot Investigation. Front. Immunol. 2023, 14, 1114239. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Eghbali, M. Influence of Sex Differences on microRNA Gene Regulation in Disease. Biol. Sex Differ. 2014, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, I.; Dejager, L.; Libert, C. X-Chromosome-Located microRNAs in Immunity: Might They Explain Male/Female Differences? The X Chromosome-Genomic Context May Affect X-Located miRNAs and Downstream Signaling, Thereby Contributing to the Enhanced Immune Response of Females. BioEssays News Rev. Mol. Cell. Dev. Biol. 2011, 33, 791–802. [Google Scholar] [CrossRef]
- Cui, C.; Yang, W.; Shi, J.; Zhou, Y.; Yang, J.; Cui, Q.; Zhou, Y. Identification and Analysis of Human Sex-Biased MicroRNAs. Genom. Proteom. Bioinform. 2018, 16, 200–211. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, X.; Xu, B.; Wang, Z.; Chen, Y.; Deng, M. Differential Expression of Plasma-derived Exosomal miRNAs in Polycystic Ovary Syndrome as a Circulating Biomarker. Biomed. Rep. 2023, 19, 92. [Google Scholar] [CrossRef]
- Zhong, S.; Borlak, J. Sex Differences in the Tumor Promoting Effects of Tobacco Smoke in a cRaf Transgenic Lung Cancer Disease Model. Arch. Toxicol. 2024, 98, 957–983. [Google Scholar] [CrossRef]
- Malmhäll, C.; Weidner, J.; Rådinger, M. MicroRNA-155 Expression Suggests a Sex Disparity in Innate Lymphoid Cells at the Single-Cell Level. Cell. Mol. Immunol. 2020, 17, 544–546. [Google Scholar] [CrossRef]
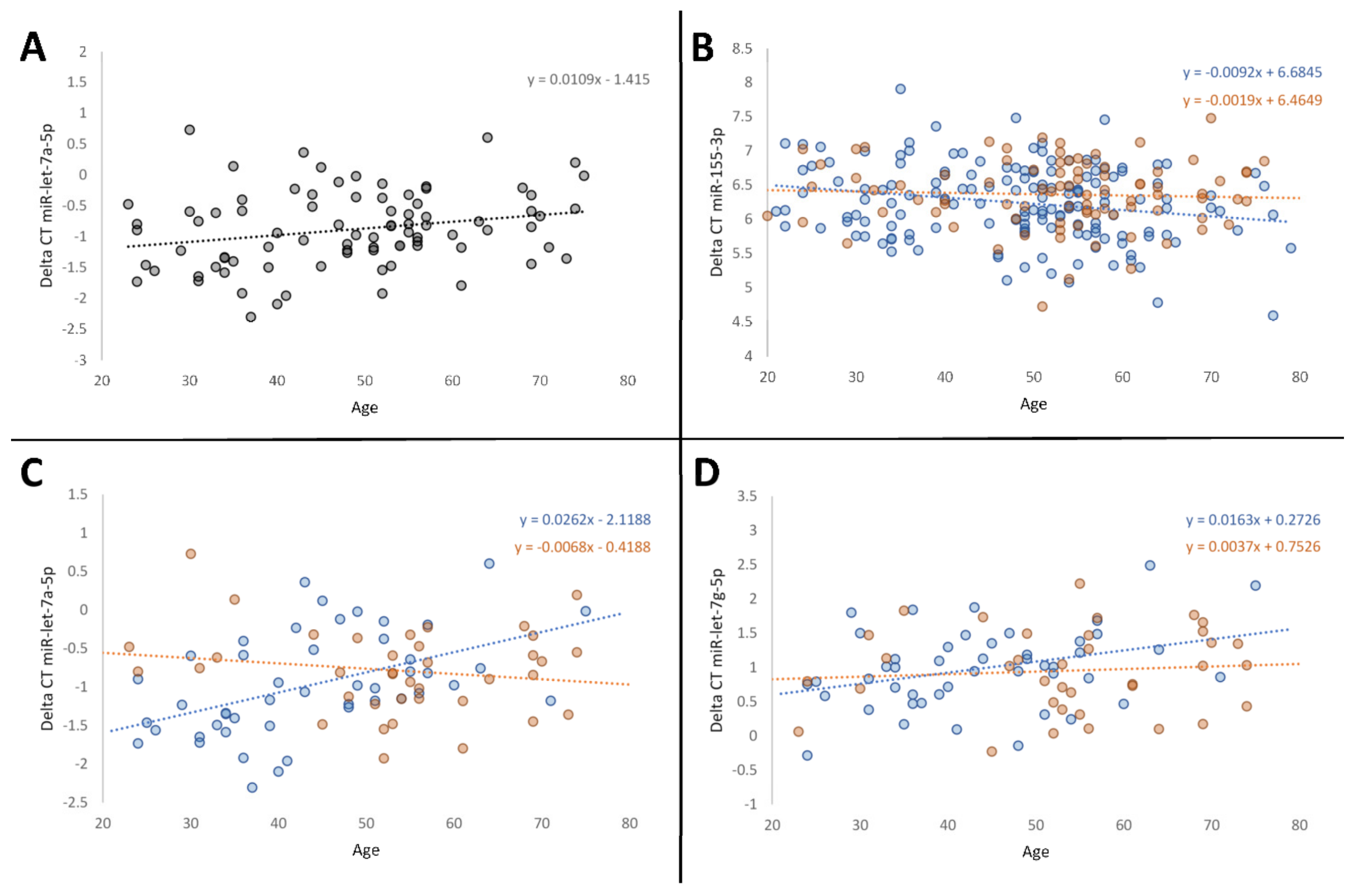
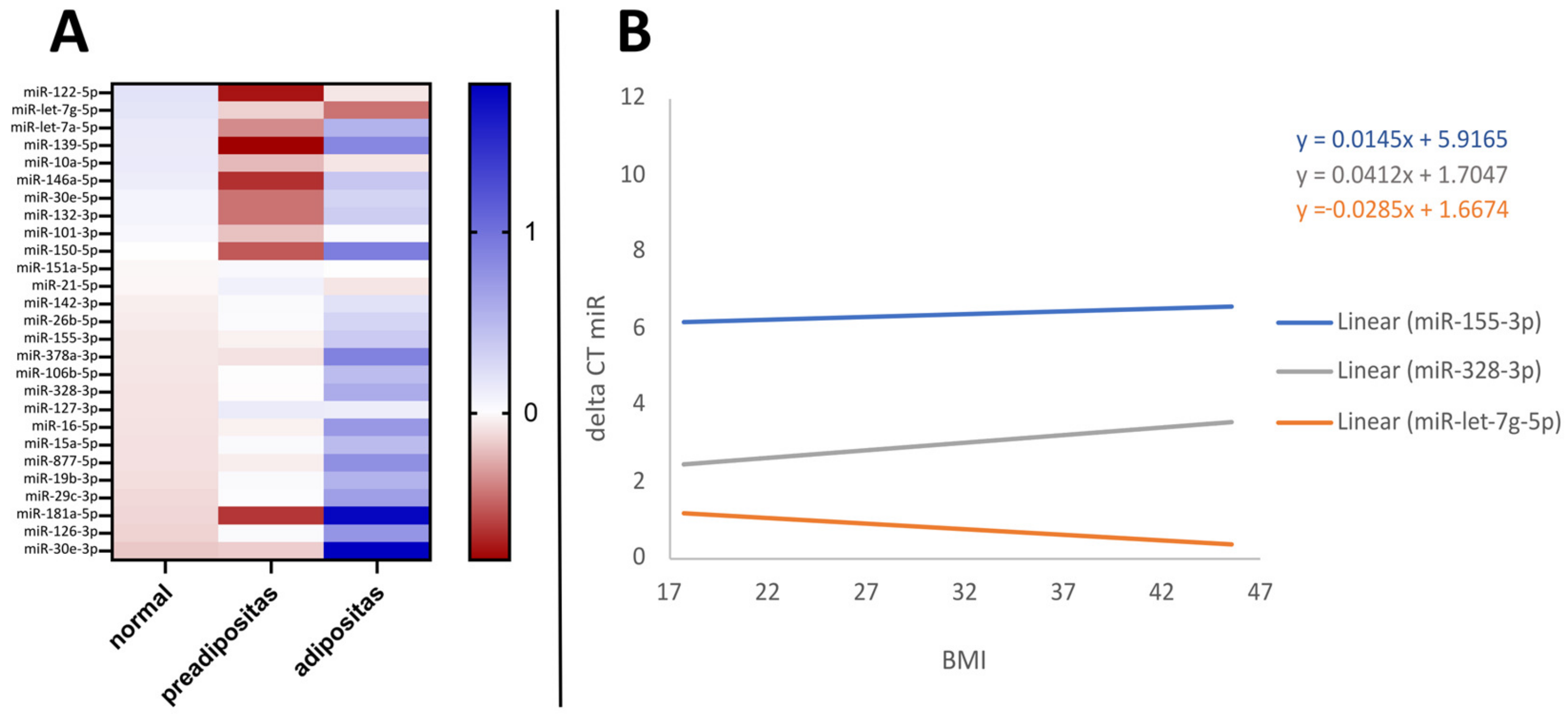
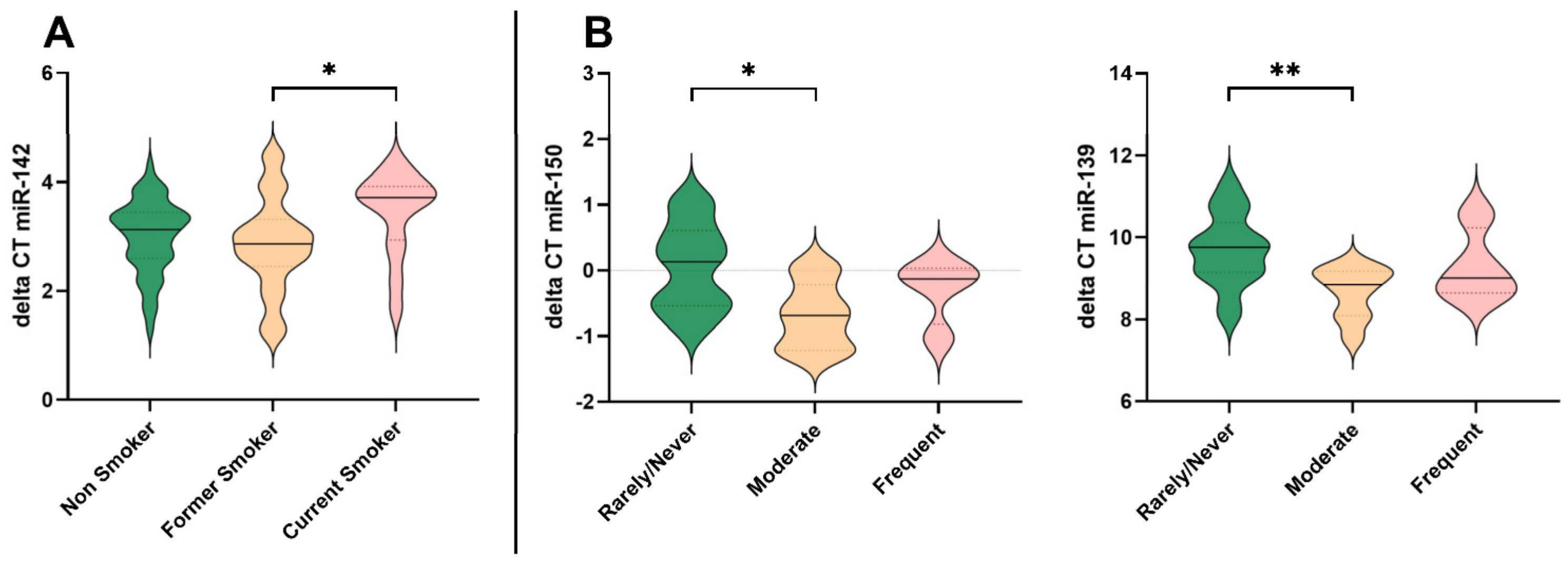
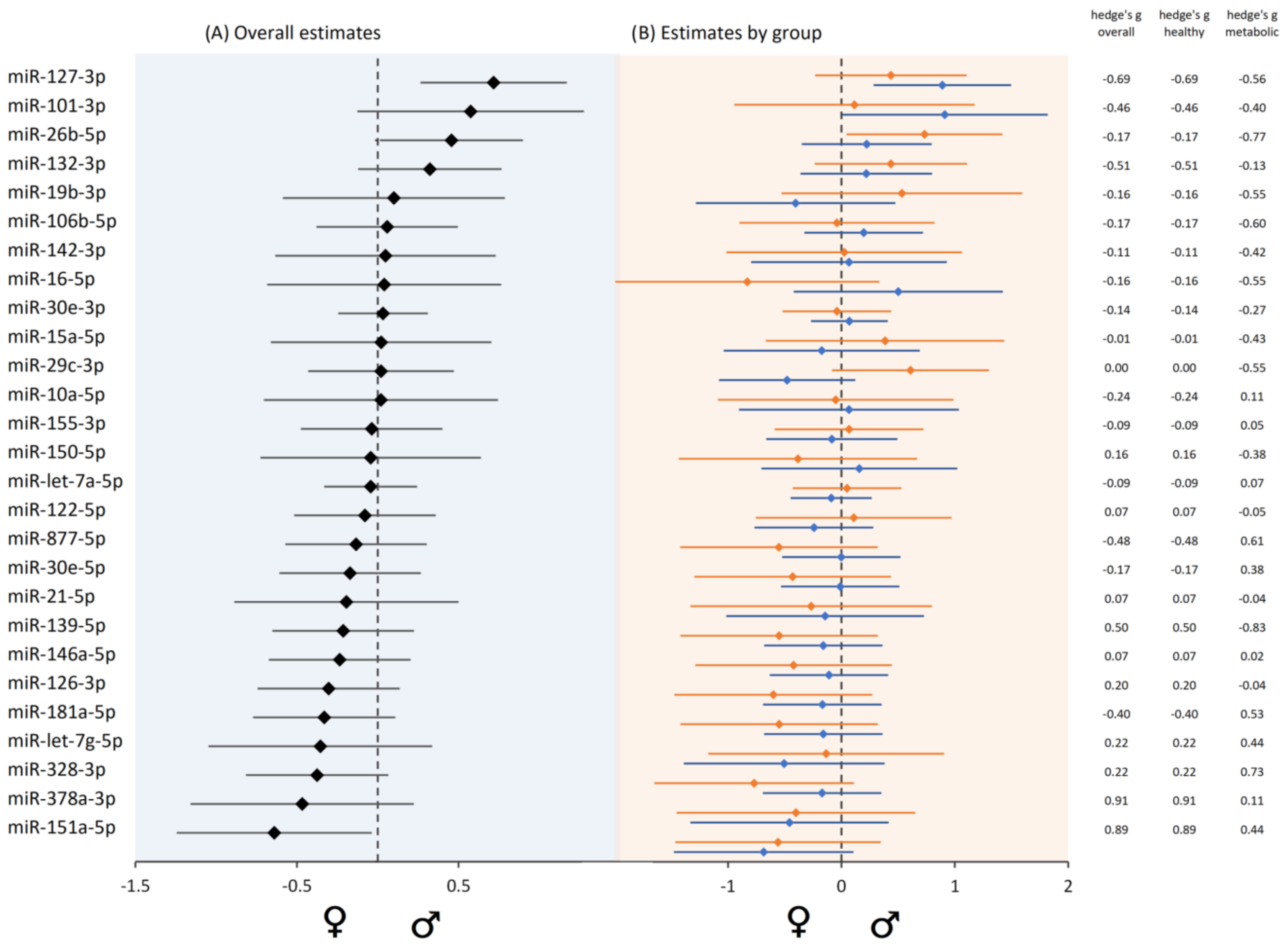

| Study Population | N | Age ± SD (Years) | Age Range (Years) | BMI ± SD (kg/m2) | BMI Range (kg/m2) | Female (N) | Male (N) |
|---|---|---|---|---|---|---|---|
| Healthy | 197 | 49.17 ± 13.18 | 21–80 | 23.67 ± 3.02 | 17.72–29.90 | 130 | 67 |
| Metabolic disease | 108 | 53.59 ± 13.78 | 20–92 | 28.20 ± 6.14 | 16.37–45.55 | 79 | 29 |
| Thyroid dysfunction | 48 | 54.21 ± 13.68 | 20–92 | 24.60 ± 4.34 | 16.37–37.13 | 44 | 4 |
| Obesity | 37 | 50.19 ± 13.65 | 23–76 | 33.59 ± 3.76 | 30.08–45.55 | 25 | 12 |
| Type 2 diabetes mellitus | 7 | 58.29 ± 17.54 | 24–74 | 30.24 ± 6.54 | 24.74–43.51 | 3 | 4 |
| Type 1 diabetes mellitus | 4 | 51.25 ± 16.34 | 35–69 | 28.14 ± 6.59 | 21.87–37.18 | 1 | 3 |
| Gout | 4 | 58.75 ± 16.28 | 47–82 | 22.98 ± 5.84 | 18.56–30.86 | 2 | 2 |
| Other | 4 | 61.00 ± 2.45 | 58–64 | 27.15 ± 7.53 | 21.63–38.22 | 2 | 2 |
| Lipid metabolism dysfunction | 3 | 50.67 ± 12.22 | 40–64 | 23.06 ± 4.30 | 19.36–27.77 | 2 | 1 |
| Glucose intolerance | 1 | 55 | 20–92 | 28.29 | 44 | 1 | |
| Whole sample group | 305 | 50.74 ± 13.54 | 20–92 | 25.27 ± 4.88 | 16.37–45.55 | 209 | 96 |
| Smoking Behaviour | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Whole Sample Group | Healthy | Metabolic Disease | |||||||
| delta CT miR-142 | no | former | daily | no | former | daily | no | former | daily |
| M | 3.009 | 2.857 | 3.446 | 3.010 | 2.603 | 3.397 | 3.006 | 3.131 | 3.535 |
| (SD) | (0.678) | (0.865) | (0.771) | (0.723) | (0.860) | (0.631) | (0.581) | (0.814) | (1.059) |
| Madj | 2.998 | 2.818 | 3.546 | 2.882 | 2.390 | 3.510 | 2.925 | 3.154 | 3.775 |
| (SE) | (0.153) | (0.154) | (0.215) | (0.263) | (0.222) | (0.275) | (0.276) | (0.296) | (0.472) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pointner, A.; Krammer, U.D.B.; Tomeva, E.; Magnet, U.; Hippe, B.; Jacob, U.; Haslberger, A.G. Lifestyle-Driven Variations in Nutrimiromic MicroRNA Expression Patterns across and beyond Genders. Life 2024, 14, 390. https://doi.org/10.3390/life14030390
Pointner A, Krammer UDB, Tomeva E, Magnet U, Hippe B, Jacob U, Haslberger AG. Lifestyle-Driven Variations in Nutrimiromic MicroRNA Expression Patterns across and beyond Genders. Life. 2024; 14(3):390. https://doi.org/10.3390/life14030390
Chicago/Turabian StylePointner, Angelika, Ulrike D. B. Krammer, Elena Tomeva, Ulrich Magnet, Berit Hippe, Ursula Jacob, and Alexander G. Haslberger. 2024. "Lifestyle-Driven Variations in Nutrimiromic MicroRNA Expression Patterns across and beyond Genders" Life 14, no. 3: 390. https://doi.org/10.3390/life14030390







