In Silico and In Vitro Studies of Novel Azomethines on DNA Repair Genes in Gastric Cell Lines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis Procedure
2.2. In Silico Calculations
2.3. Cell Culture
2.4. In Vitro Cytotoxicity Determination (MTT)
2.5. Cell Morphology
2.6. Bioinformatics Analysis
2.7. RNA Isolation from Cell Culture Samples
2.8. cDNA Synthesis
2.9. Real-Time PCR Analysis
2.10. Determination of Antioxidant Levels
2.10.1. Determination of Glutathione (GSH)
2.10.2. Determination of Glutathione-S-Transferase (GST)
2.10.3. Catalase Determination (CAT)
2.11. Membrane Integrity
Determination of Lactate Dehydrogenase (LDH)
3. Results
3.1. Synthesis Part
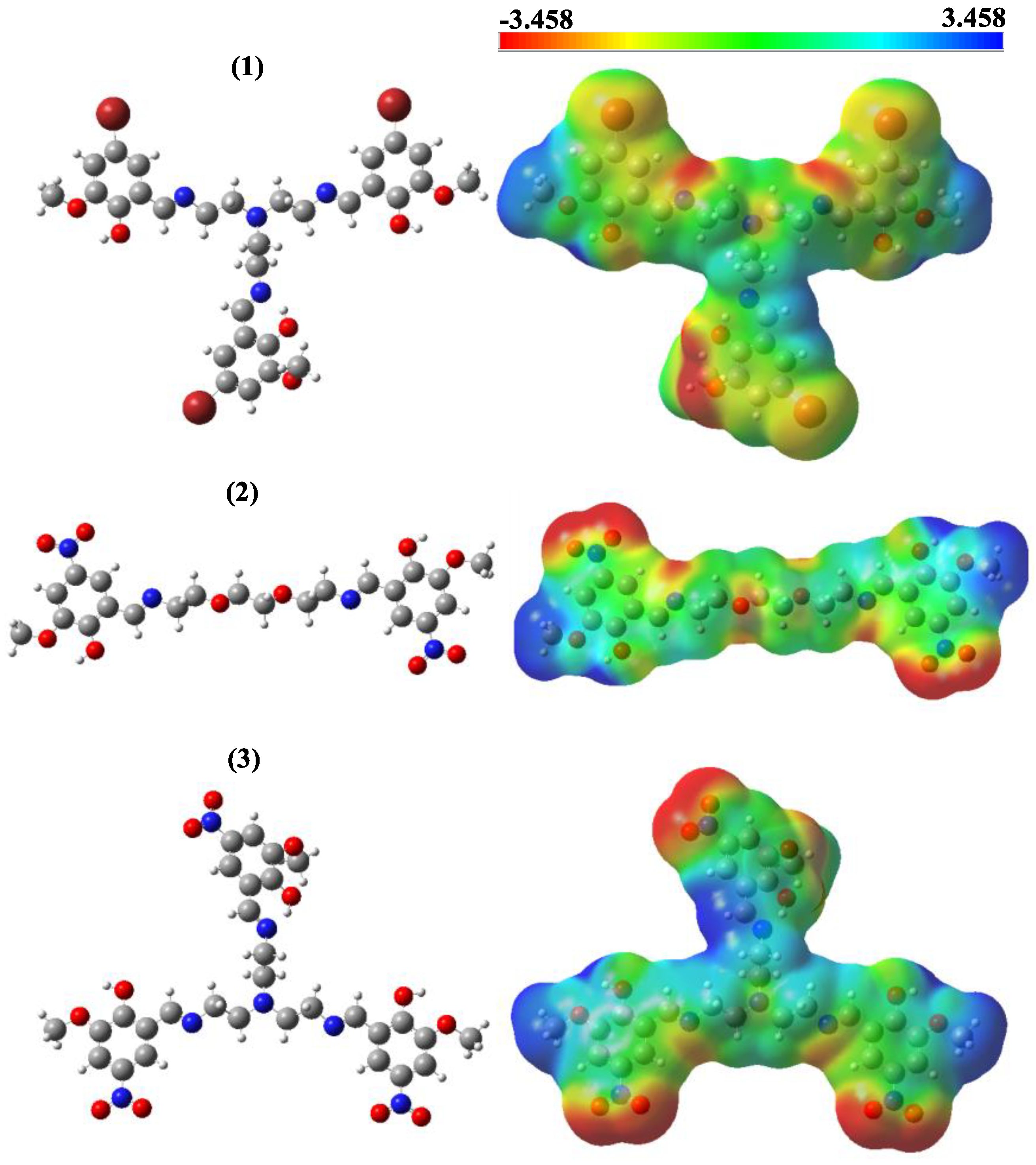
3.2. In Silico Studies
Frontier Molecular Orbitals (FMOs) and MEP Contours
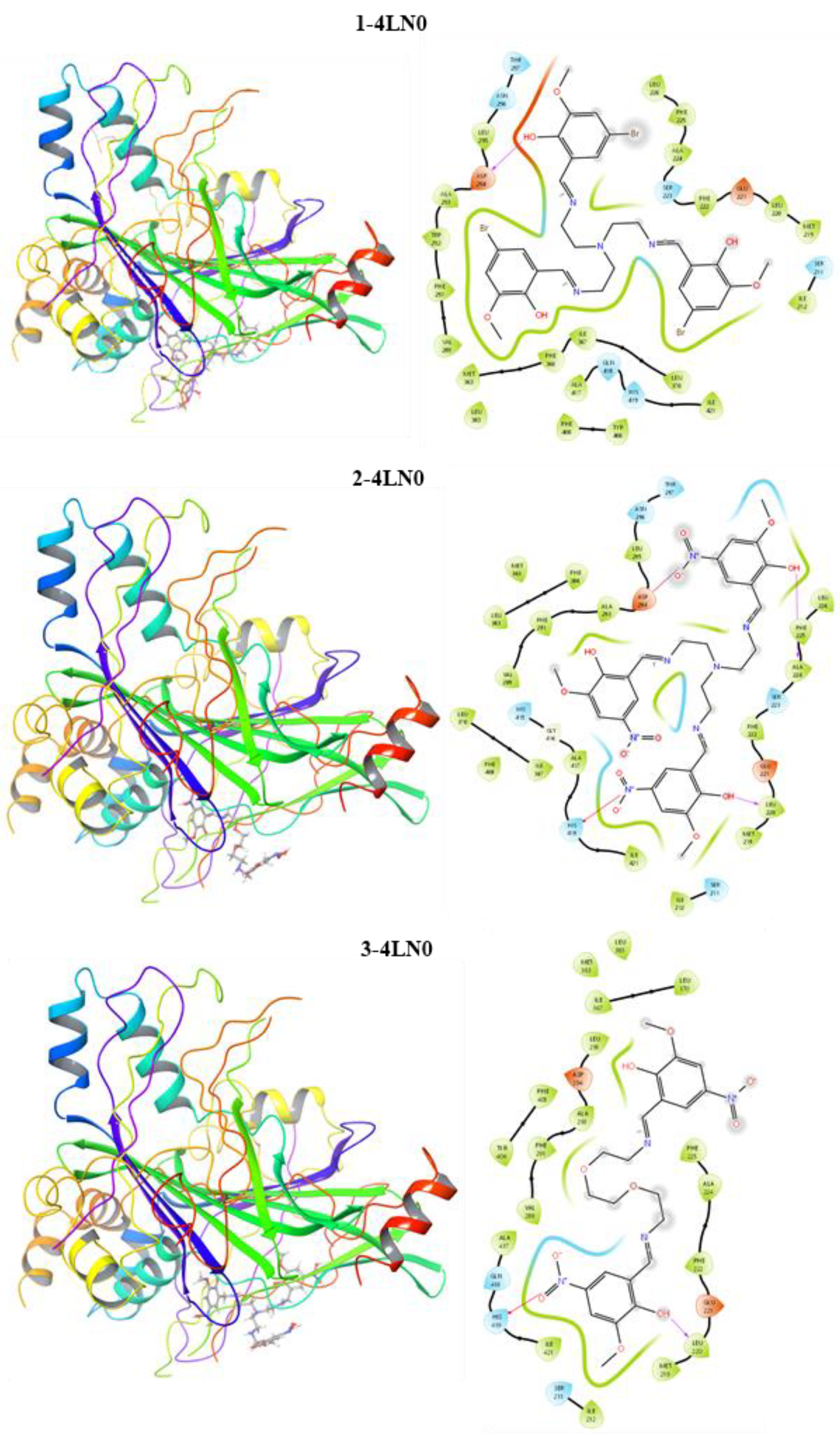
3.3. Molecular Docking
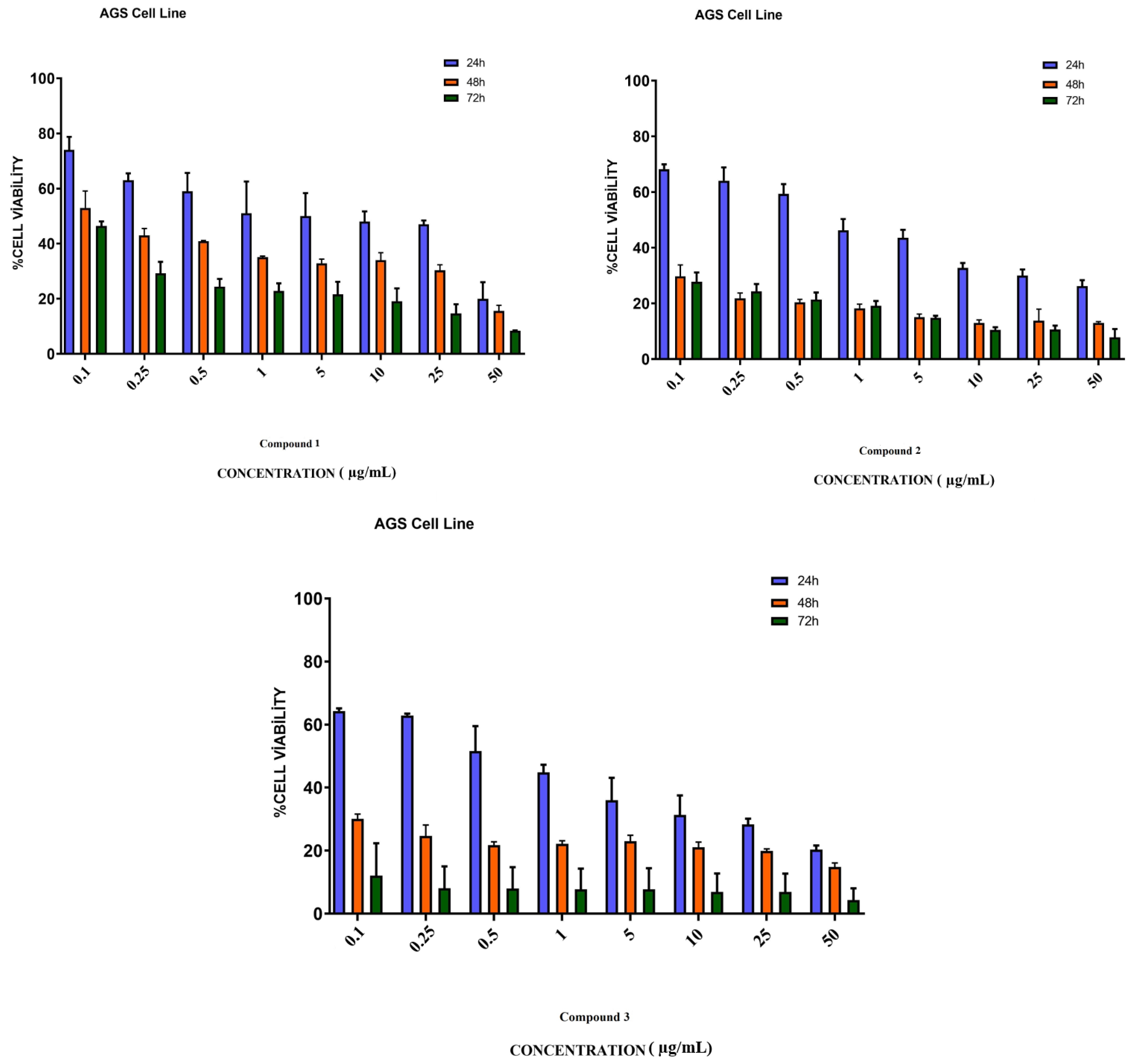
3.4. In Vitro Assay for Cytotoxicity Activity (MTT Assay)
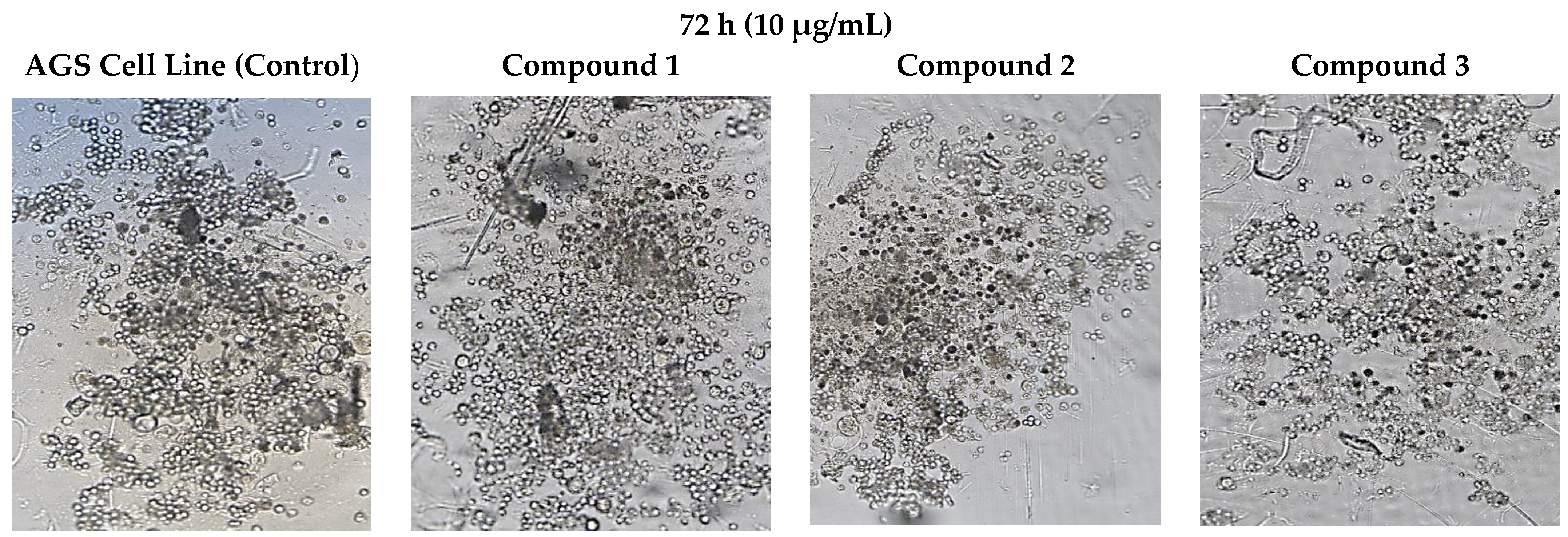
3.5. Cell Morphology Analysis
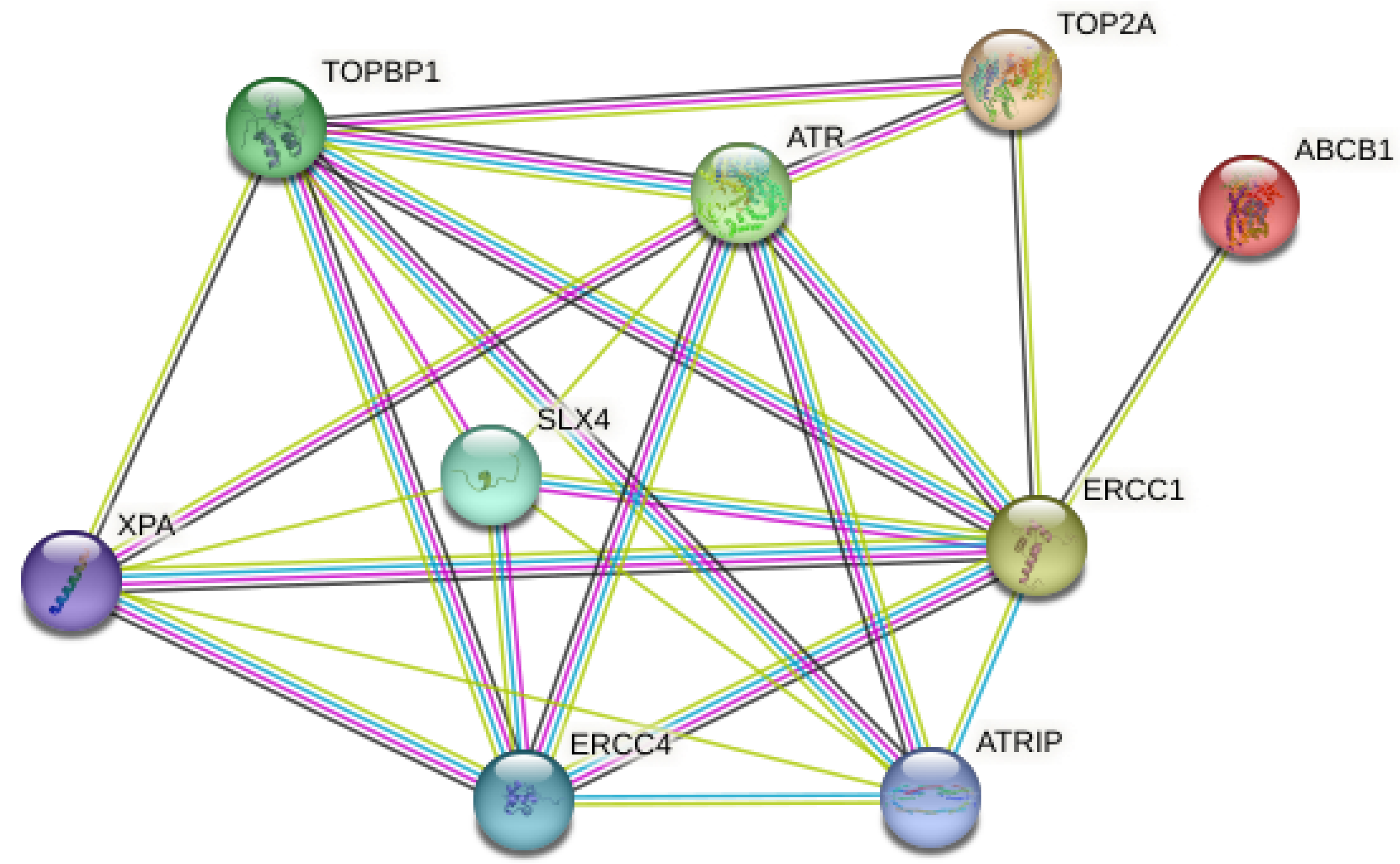
3.6. Bioinformatics Analysis
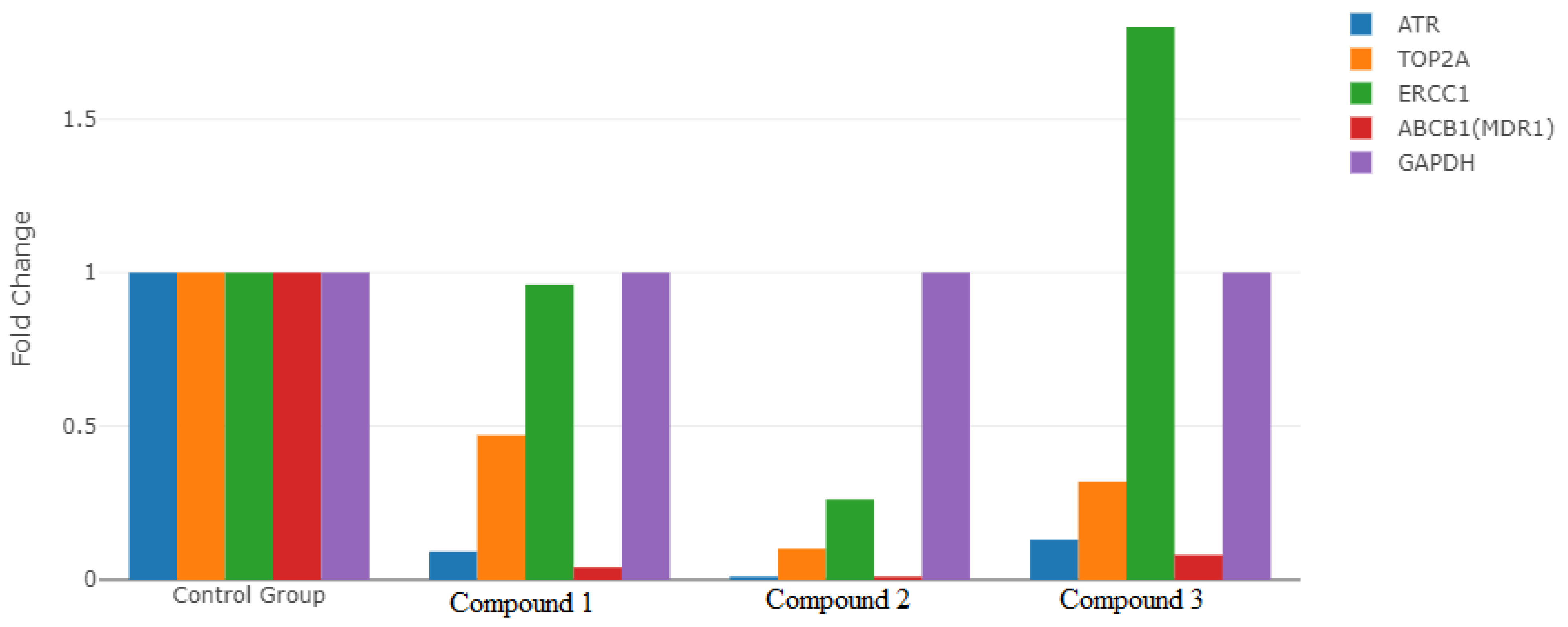
3.7. Gene Expression Analysis
3.8. Determination of Antioxidant Levels
3.8.1. Glutathione Determination (GSH)
3.8.2. Glutathione-S-Transferase Determination (GST)
3.8.3. Catalase Determination (CAT)
3.9. Membrane Integrity
Lactate Dehydrogenase Determination
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ji, B.T.; Chow, W.H.; Yang, G.; McLaughlin, J.K.; Gao, R.N.; Zheng, W.; Shu, X.O.; Jin, F.; Fraumeni, J.F., Jr.; Gao, Y.T. The influence of cigarette smoking, alcohol, and green tea consumption on the risk of carcinoma of the cardia and distal stomach in Shanghai, China. Cancer 1996, 77, 2449–2457. [Google Scholar] [CrossRef]
- Catalano, V.; Labianca, R.; Beretta, G.D.; Gatta, G.; de Braud, F.; Van Cutsem, E. Gastric cancer. Crit. Rev. Oncol. Hematol. 2009, 71, 127–164. [Google Scholar] [CrossRef] [PubMed]
- Houghton, J.; Wang, T.C. Helicobacter pylori and gastric cancer: A new paradigm for inflammation-associated epithelial cancers. Gastroenterology 2005, 128, 1567–1578. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.K.; Cho, C.H.; Lee, C.W.; Fan, D.; Wu, K.; Yu, J.; Sung, J.J. Dysregulation of cellular signaling in gastric cancer. Cancer Lett. 2010, 295, 144–153. [Google Scholar] [CrossRef]
- Yamashita, K.; Sakuramoto, S.; Watanabe, M. Genomic and epigenetic profiles of gastric cancer: Potential diagnostic and therapeutic applications. Surg. Today 2011, 41, 24–38. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J.; Siggens, L.; Figg, N.; Bennett, M.; Foo, R.; Chastain, P.D.; Nakamura, J.; et al. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef]
- Sancar, A.; Lindsey-Boltz, L.A.; Unsal-Kaçmaz, K.; Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004, 73, 39–85. [Google Scholar] [CrossRef]
- Thomas, S.D. DNA Repair Pathways and Mechanisms. In DNA Repair of Cancer Stem Cells; Springer: Dordrecht, Germany, 2012; pp. 19–32. [Google Scholar] [CrossRef]
- Gossage, L.; Perry, C.; Abbotts, R.; Madhusudan, S.B. Base excision repair factors are promising prognostic and predictive markers in cancer. Curr. Mol. Pharmacol. 2012, 5, 115–124. [Google Scholar] [CrossRef]
- Kakde, D.; Jain, D.; Shrivastava, V.; Kakde, R.; Patil, A.T. Cancer Therapeutics-Opportunities, Challenges and Advances in Drug Delivery. J. Appl. Pharm. Sci. 2011, 9, 1–10. [Google Scholar]
- Sarigul, M.; Sari, A.; Kose, M.; McKee, V.; Elmastas, M.; Demirtas, I.; Kurtoglu, M. New bioactive azoazomethine based Cu(II) complexes. Inorganica Chim. Acta 2016, 444, 166–175. [Google Scholar] [CrossRef]
- Hassan, A.S.; Askar, A.A.; Nossier, E.S.; Naglah, A.M.; Moustafa, G.O.; Al-Omar, M.A. Antibacterial evaluation, in silico characters and molecular docking of Schiff bases derived from 5-aminopyrazoles. Molecules 2019, 24, 3130. [Google Scholar] [CrossRef]
- Al-Shareef, H.F.; Elhady, H.A.; Aboellil, A.H.; Hussein, E.M. Ammonium chloride catalyzed synthesis of novel Schiff bases from spiro[indoline-3,4′-pyran]-3′-carbonitriles and evaluation of their antimicrobial and anti-breast cancer activities. Springerplus 2016, 5, 887. [Google Scholar] [CrossRef] [PubMed]
- Hussein, E.M.; Masaret, G.S.; Khairou, K.S. Efficient synthesis and antimicrobial evaluation of some Mannich bases from 2-arylidine-1-thia-4-azaspiro[4.5]decan-3-ones. Chem. Cent. J. 2015, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Kose, A.; Erkan, S.; Tümer, M. A series of phenanthroline-imine compounds: Computational, OLED properties and fluorimetric sensing of nitroaromatic compounds. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2023, 286, 122006. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Wang, H.; Shi, Z.; Dong, A.; Zhang, W.; Song, X.; He, F.; Wang, Y.; Zhang, Z.; Wang, W. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 2014, 25, 166–180. [Google Scholar] [CrossRef]
- Giustarini, D.; Dalle-Donne, I.; Milzani, A.; Fanti, P.; Rossi, R. Analysis of GSH and GSSG afterderivatizationwith N-ethylmaleimide. Nat. Protoc. 2013, 8, 1660–1669. [Google Scholar] [CrossRef]
- Ghelfi, A.; Gaziola, S.A.; Cia, M.C.; Chabregas, S.M.; Falco, M.C.; Kuser-Falcão, P.R.; Azevedo, R.A. Cloning, expression, molecular modelling and docking analysis of glutathione transferase from Saccharum officinarum. Ann. Appl. Biol. 2011, 159, 267–280. [Google Scholar] [CrossRef]
- Aebi, H. Catalase. In Methods of Enzymatic Analysis; Academic Press: Cambridge, MA, USA, 1974; pp. 673–684. [Google Scholar]
- Decker, T.; Lohmann-Matthes, M.L. A quick and simple method for the quantitation of lactate dehydrogenase release in measurements of cellular cytotoxicity and tumor necrosis factor (TNF) activity. J. Immunol. Methods 1988, 115, 61–69. [Google Scholar] [CrossRef]
- Kaya, S.; Erkan, S.; Karakaş, D. Computational investigation of molecular structures, spectroscopic properties and antitumor-antibacterial activities of some Schiff bases. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 244, 118829. [Google Scholar] [CrossRef]
- Kaya, S.; Erkan, S.; Karakaş, D. Computational design and characterization of platinum-II complexes of some Schiff bases and investigation of their anticancer–antibacterial properties. Appl. Organomet. Chem. 2022, 36, e6805. [Google Scholar] [CrossRef]
- Sayin, K.; Üngördü, A. Investigations of structural, spectral and electronic properties of enrofloxacin and boron complexes via quantum chemical calculation and molecular docking. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2019, 220, 117102. [Google Scholar] [CrossRef] [PubMed]
- Yakan, H.; Koçyiğit, Ü.M.; Muğlu, H.; Ergul, M.; Erkan, S.; Güzel, E.; Gülçin, İ. Potential thiosemicarbazone-based enzyme inhibitors: Assessment of antiproliferative activity, metabolic enzyme inhibition properties, and molecular docking calculations. J. Biochem. Mol. Toxicol. 2022, 36, e23018. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.J.; Rillema, D.P.; Basolo, F.J. Oxygen carrier and redox properties of some neutral cobalt chelates. Axialand in-plane ligand effects. Am. Chem. Soc. 1974, 96, 392–400. [Google Scholar] [CrossRef]
- Asada, H.; Hayashi, K.; Negoro, S.; Fujiwara, M.; Matsushita, T. Preparation andstructures of trinuclear manganese (II) complexes with N-2-pyridiylmethylidene-2-hydroxy-5-substituted phenylamine. Inorg. Chem. Commun. 2003, 6, 193–196. [Google Scholar] [CrossRef]
- Saghatforoush, L.; Aminkhani, A.; Ershad, S.; Karimnezhad, G.; Ghammamy, S.; Kabiri, R. Preparation of Zinc (II) and cadmium (II) complexes of the tetradentate Schiff base ligand 2-((E)-(2-(2-(pyridine-2-yl)- ethylthio)ethylimino)methyl)-4-bromophenol (PytBrsalH). Molecules 2008, 13, 804–811. [Google Scholar] [CrossRef]
- Chen, W.; Ou, W.; Wang, L.; Hao, Y.; Cheng, J.; Li, J.; Liu, Y.N. Synthesis and biological evaluation of hydroxyl-substituted Schiff-bases containing ferrocenyl moieties. Dalton Trans. 2013, 42, 15678–15686. [Google Scholar] [CrossRef]
- Cheng, L.X.; Tang, J.J.; Luo, H.; Jin, X.L.; Dai, F.; Yang, J.; Qian, Y.P.; Li, X.Z.; Zhou, B. Antioxidant and antiproliferative activities of hydroxyl-substituted Schiff bases. Bioorg Med. Chem. Lett. 2010, 20, 2417–2420. [Google Scholar] [CrossRef]
- Vinodkumar, J.K.; Pushpavathi, I.; Keerthikumar, J.T.; Maliyappa, M.R.; Ravi, B.N. Synthesis, characterization, computational and biological studies of nitrothiazole incorporated heterocyclic azo dyes. Struct. Chem. 2020, 31, 37. [Google Scholar] [CrossRef]
- Vinodkumar, J.K. Synthesis, structural investigations and in vitro biological evaluation of N, N-dimethyl aniline derivatives based azo dyes as potential pharmacological agents. J. Mol. Struct. 2019, 1186, 404–412. [Google Scholar] [CrossRef]
- Erdoğan, M.; Yıldız, B.; Tüzün, B.; Özden, Ö. Synthesis and Characterization of Some Benzidine-Based Azomethine Derivatives with Molecular Docking Studies and Anticancer Activities. Chem. Pap. 2023, 77, 6829–6847. [Google Scholar] [CrossRef]
- Bouwman, P.; Jonkers, J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat. Rev. Cancer 2012, 12, 587–598. [Google Scholar] [CrossRef]
- LaTulippe, E.; Satagopan, J.; Smith, A.; Scher, H.; Scardino, P.; Reuter, V.; Gerald, W.L. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002, 62, 4499–4506. [Google Scholar]
- Wei, Q.; Cheng, L.; Xie, K.; Bucana, C.D.; Dong, Z. Direct correlation between DNA repair capacity and metastatic potential of K-1735 murine melanoma cells. J. Investig. Dermatol. 1997, 108, 3–6. [Google Scholar] [CrossRef]
- Sarasin, A.; Kauffmann, A. Overexpression of DNA repair genes is associated with metastasis: A new hypothesis. Mutat. Res. Rev. Mutat. Res. 2008, 659, 49–55. [Google Scholar] [CrossRef]
- Taniguchi, T.; Tischkowitz, M.; Ameziane, N.; Hodgson, S.V.; Mathew, C.G.; Joenje, H.; D’Andrea, A.D. Disruption of the Fanconi anemia—BRCA pathway in cisplatin sensitive ovarian tumors. Nat. Med. 2003, 9, 568–574. [Google Scholar] [CrossRef]
- Tanaka, A.; Weinel, S.; Nagy, N.; O’Driscoll, M.; Lai-Cheong, J.E.; Kulp-Shorten, C.L.; Knable, A.; Carpenter, G.; Fisher, S.A.; Hiragun, M. Germline mutation in ATR in autosomal-dominant oropharyngeal cancer syndrome. Am. J. Hum. Genet. 2012, 90, 511–517. [Google Scholar] [CrossRef]
- Helt, C.E.; Cliby, W.A.; Keng, P.C.; Bambara, R.A.; O’Reilly, M.A. Ataxia telangiectasia mutated (ATM) and ATM and Rad3-related protein exhibit selective target specificities in response to different forms of DNA damage. J. Biol. Chem. 2005, 280, 1186–1192. [Google Scholar] [CrossRef]
- Wang, X.; Khadpe, J.; Hu, B.; Iliakis, G.; Wang, Y. An overactivated ATR/CHK1 pathway is responsiblefortheprolonged G2 accumulation in irradiated AT cells. J. Biol. Chem. 2003, 278, 30869–30874. [Google Scholar] [CrossRef]
- Parikh, R.A.; Appleman, L.J.; Bauman, J.E.; Sankunny, M.; Lewis, D.W.; Vlad, A.; Gollin, S.M. Upregulation of the ATR-CHEK1 pathway in oral squamous cell carcinomas. Genes Chromosomes Cancer 2014, 53, 25–37. [Google Scholar] [CrossRef]
- Gregg, S.Q.; Robinson, A.R.; Niedernhofer, L.J. Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease. DNA Repair 2011, 10, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Kashiyama, K.; Nakazawa, Y.; Pilz, D.T.; Guo, C.; Shimada, M.; Sasaki, K.; Fawcett, H.; Wing, J.F.; Lewin, S.O.; Carr, L.; et al. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am. J. Hum. Genet. 2013, 92, 807–819. [Google Scholar] [CrossRef] [PubMed]
- Olaussen, K.A.; Mountzios, G.; Soria, J.C. ERCC1 as a risk stratifier in platinum-based chemotherapy for nonsmall-cell lung cancer. Curr. Opin. Pulm. Med. 2007, 13, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Simon, G.R.; Sharma, S.; Cantor, A.; Smith, P.; Bepler, G. ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest 2005, 127, 978–983. [Google Scholar] [CrossRef]
- Sng, J.H.; Heaton, V.J.; Bell, M.; Maini, P.; Austin, C.A.; Fisher, L.M. Molecular cloning and characterization of the human topoisomerase II alpha and II beta genes: Evidence for isoform evolution through gene duplication. Biochim. Biophys. Acta 1999, 1444, 395–406. [Google Scholar] [CrossRef]
- Smith, K.; Houlbrook, S.; Greenall, M.; Carmichael, J.; Harris, A.L. Topoisomerase II alpha co-amplification with erbB2 in human primary breast cancer and breast cancer cell lines: Relationship to m-AMSA andmitoxantrone sensitivity. Oncogene 1993, 8, 933–938. [Google Scholar]
- Mueller, R.E.; Parkes, R.K.; Andrulis, I.; O’Malley, F.P. Amplification of the TOP2A gene does not predict high levels of topoisomerase II alpha protein in human breast tumor samples. Genes Chromosomes Cancer 2004, 39, 288–297. [Google Scholar] [CrossRef]
- Järvinen, T.A.; Tanner, M.; Rantanen, V.; Barlund, M.; Borg, A.; Grenman, S.; Isola, J. Amplification and deletion of topoisomerase IIalpha associate with ErbB-2 amplification and affect sensitivity to topoisomerase II inhibitor doxorubicin in breast cancer. Am. J. Pathol. 2000, 156, 839–847. [Google Scholar] [CrossRef]
- De Resende, M.F.; Vieira, S.; Chinen, L.T.D.; Chiappelli, F.; da Fonseca, F.P.; Guimarães, G.C.; Rocha, R.M. Prognostication of prostate cancer based on TOP2A protein and gene assessment: TOP2A in prostate cancer. J. Transl. Med. 2013, 11, 36. [Google Scholar] [CrossRef]
- Mizutani, T.; Masuda, M.; Nakai, E.; Furumiya, K.; Togawa, H.; Nakamura, Y.; Kawai, Y.; Nakahira, K.; Shinkai, S.; Takahashi, K. Genuine functions of P-glycoprotein (ABCB1). Curr. Drug Metab. 2008, 9, 167–174. [Google Scholar] [CrossRef]
- Seo, J.; Lee, C.R.; Paeng, J.C.; Kwon, H.W.; Lee, D.; Kim, S.C.; Han, J.; Ku, J.L.; Chae, J.H.; Lim, B.C.; et al. Biallelic mutations in ABCB1 display recurrent reversible encephalopathy. Ann. Clin. Transl. Neurol. 2020, 7, 1443–1449. [Google Scholar] [CrossRef]
- Haenisch, S.; Zimmermann, U.; Dazert, E.; Wruck, C.J.; Dazert, P.; Siegmund, W.; Kroemer, H.K.; Warzok, R.W.; Cascorbi, I. Influence of polymorphisms of ABCB1 and ABCC2 on mRNA and protein expression in normal andcancerouskidneycortex. Pharm. J. 2007, 7, 56–65. [Google Scholar] [CrossRef]
- De, I.S.; De, P.A.; Stocco, G.; Bartoli, F.; Bussani, R.; Decorti, G. ABCB1 gene polymorphisms and expression of P-glycoprotein and long-term prognosis in colorectal cancer. Anticancer Res. 2008, 28, 3921–3928. [Google Scholar]
- Linn, S.C.; Giaccone, G. MDR1/P-glycoprotein expression in colorectal cancer. Eur. J. Cancer 1995, 31, 1291–1294. [Google Scholar] [CrossRef] [PubMed]
- Gamcsik, M.P.; Kasibhatla, M.S.; Teeter, S.D.; Colvin, O.M. Glutathione levels in human tumors. Biomarkers 2012, 17, 671–691. [Google Scholar] [CrossRef] [PubMed]
- Harris, I.S.; Treloar, A.E.; Inoue, S.; Sasaki, M.; Gorrini, C.; Lee, K.C.; Yung, K.Y.; Brenner, D.; Knobbe-Thomsen, C.B.; Cox, M.A.; et al. Glutathione and thioredox in antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell 2015, 27, 211–222. [Google Scholar] [CrossRef]
- Cramer, S.L.; Saha, A.; Liu, J.; Tadi, S.; Tiziani, S.; Yan, W.; Triplett, K.; Lamb, C.; Alters, S.E.; Rowlinson, S.; et al. Systemic depletion of L-cyst(e)ine with cyst(e)inase increases reactive oxygen species and suppresses tumor growth. Nat. Med. 2017, 23, 120–127. [Google Scholar] [CrossRef]
- Mitchell, J.B.; Russo, A. The role of glutathione in radiation and drug induced cytotoxicity. Br. J. Cancer Suppl. 1987, 8, 96–104. [Google Scholar]
- Estrela, J.M.; Ortega, A.; Obrador, E. Glutathione in cancer biology and therapy. Crit. Rev. Clin. Lab. Sci. 2006, 43, 143–181. [Google Scholar] [CrossRef]
- Balendiran, G.K.; Dabur, R.; Fraser, D. The role of glutathione in cancer. Cell Biochem. Funct. 2004, 22, 343–352. [Google Scholar] [CrossRef]
- Sherratt, P.J.; Hayes, J.D. Glutathione S-transferases. In Enzyme Systems That Metabolise Drugs and Other Xenobiotics; Ioannides, C., Ed.; Wiley: Chichester, UK, 2001; pp. 319–352. [Google Scholar]
- Rowe, J.D.; Nieves, E.; Listowsky, I. Subunit diversity and tissue distribution of human glutathione S-transferases: Interpretations based on electrosprayionization-MS and peptide sequence-specific antisera. Biochem. J. 1997, 325, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Gupta, S. The multifaceted role of glutathione S-transferases in cancer. Cancer Lett. 2018, 433, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Moloney, J.N.; Cotter, T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar] [CrossRef]
- VNogueira, V.; Park, Y.; Chen, C.C.; Xu, P.Z.; Chen, M.L.; Tonic, I.; Unterman, T.; Hay, N. Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell 2008, 14, 458–470. [Google Scholar] [CrossRef] [PubMed]
- Trachootham, D.; Zhou, Y.; Zhang, H.; Demizu, Y.; Chen, Z.; Pelicano, H.; Chiao, P.J.; Achanta, G.; Arlinghaus, R.B.; Liu, J.; et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell 2006, 10, 241–252. [Google Scholar] [CrossRef]
- Handschuh, L.; Kazmierczak, M.; Milewski, M.C.; Góralski, M.; Łuczak, M.; Wojtasz Wska, M.; Uszczyńska-Ratajczak, B.; Lewandowski, K.; Komarnicki, M.; Figlerowicz, M. Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1 and -M2 through boutique microarrays, real-time PCR and droplet digital PCR. Int. J. Oncol. 2018, 52, 656–678. [Google Scholar] [CrossRef]
- Augoff, K.; Hryniewicz-Jankowska, A.; Tabola, R. Lactate dehydrogenase 5: An old friend and a new hope in the war on cancer. Cancer Lett. 2015, 358, 1–7. [Google Scholar] [CrossRef]
- Gallo, M.; Sapio, L.; Spina, A.; Naviglio, D.; Calogero, A.; Naviglio, S. Lactic dehydrogenase and cancer: An overview. Front. Biosci. (Landmark Ed.) 2015, 20, 1234–1249. [Google Scholar] [CrossRef]
- García, R.; Hernández, J.M.; Caballero, M.D.; González, M.; Galende, J.; del Cañizo, M.C.; Vázquez, L.; San Miguel, J.F. Serum lactate dehydrogenase level as a prognostic factor in Hodgkin’s disease. Br. J. Cancer 1993, 68, 1227–1231. [Google Scholar] [CrossRef]
- Schneider, R.J.; Seibert, K.; Passe, S.; Little, C.; Gee, T.; Lee, B.J.; Miké, V.; Young, C.W. Prognostic significance of serum lactate dehydrogenase in malignant lymphoma. Cancer 1980, 46, 139–143. [Google Scholar] [CrossRef]



 | 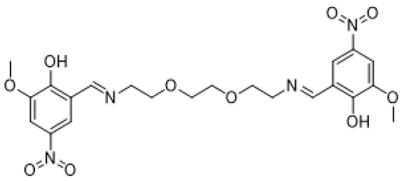 |
| ((nitrilotris(ethane-2,1-diyl))tris(azaneylylidene)) | 5,8-dioxa-2,11-diazadodeca-1,11-diene-tris(methaneylylidene))tris(4-bromo-2-methoxyphenol) |
| 1,12-diyl)bis(2-methoxy-4-nitrophenol) | |
| (Compound 1) | (Compound 2) |
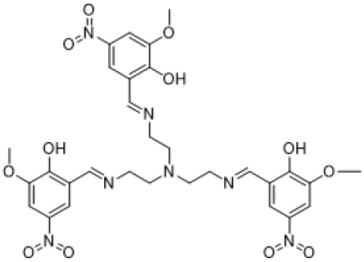 | |
| ((nitrilotris(ethane-2,1-diyl))tris(azaneylylidene))tris(methaneylylidene))tris(2-methoxy-4-nitrophenol) | |
| (Compound 3) | |
| (1)-4LN0 | (2)-4LN0 | (3)-4LN0 | Docetaxel-4LN0 | |
|---|---|---|---|---|
| DS * | −6.979 | −8.100 | −8.906 | −3.999 |
| EvdW * | −19.563 | −20.245 | −20.841 | −14.371 |
| ECoul * | −8.564 | −10.525 | −10.002 | −7.001 |
| ETotal * | −25.125 | −27.005 | −27.899 | −24.514 |
| Proteins | Associated Proteins | Predicted Functional Proteins | Homology Score |
|---|---|---|---|
| ATR | TOPBP1 | Serine/threonine protein kinase ATR | 0.999 |
| ATR | ATRIP | Serine/threonine protein kinase ATR | 0.999 |
| ATRIP | TOPBP1 | Three-prime repair exonuclease 1 | 0.999 |
| ERCC1 | SLX4 | DNA excision repair protein ERCC-1 | 0.999 |
| ERCC1 | XPA | DNA excision repair protein ERCC-1 | 0.999 |
| ERCC1 | ERCC4 | DNA excision repair protein ERCC-1 | 0.999 |
| ERCC4 | XPA | DNA repair endonuclease XPF | 0.997 |
| ERCC4 | SLX4 | DNA repair endonuclease XPF | 0.991 |
| ATR | ERCC4 | Serine/threonine protein kinase ATR | 0.986 |
| ATR | ERCC1 | Serine/threonine protein kinase ATR | 0.967 |
| TOPBP1 | ERCC4 | Topoisomerase (dna) II-binding protein 1 | 0.957 |
| ERCC1 | TOPBP1 | DNA excision repair protein ERCC-1 | 0.956 |
| ATRIP | ERCC1 | Three-prime repair exonuclease 1 | 0.941 |
| ATRIP | ERCC4 | Three-prime repair exonuclease 1 | 0.923 |
| TOP2A | TOPBP1 | DNA topoisomerase 2-alpha | 0.875 |
| XPA | ATR | DNA repair protein complementing XP-A cells | 0.801 |
| SLX4 | TOPBP1 | Structure-specific endonuclease subunit slx4 | 0.760 |
| ATR | TOP2A | Serine/threonine protein kinase ATR | 0.578 |
| ABCB1 | ERCC1 | Multidrug-resistance protein 1 | 0.530 |
| SLX4 | ATR | Structure-specific endonuclease subunit slx4 | 0.526 |
| ERCC1 | TOP2A | DNA excision repair protein ERCC-1 | 0.509 |
| XPA | SLX4 | Topoisomerase (dna) ii-binding protein 1 | 0.508 |
| TOPBP1 | XPA | Topoisomerase (dna) ii-binding protein 1 | 0.469 |
| ATRIP | XPA | DNA repair protein complementing XP-A cells | 0.462 |
| ATRIP | SLX4 | DNA repair protein complementing XP-A cells | 0.444 |
| Genes | Groups | Mean CT | Fold-Change | p-Value |
|---|---|---|---|---|
| ATR | Compound 1 | 27.29 | 0.09 | 0.001 * |
| Compound 2 | 28.89 | 0.01 | ||
| Compound 3 | 27.41 | 0.13 | ||
| Control | 27.71 | |||
| TOP2A | Compound 1 | 25.58 | 0.47 | 0.001 * |
| Compound 2 | 26.37 | 0.10 | ||
| Compound 3 | 26.78 | 0.32 | ||
| Control | 28.36 | |||
| ERCC1 | Compound 1 | 25.64 | 0.96 | 0.001 * |
| Compound 2 | 26.08 | 0.26 | ||
| Compound 3 | 25.36 | 1.80 | ||
| Control | 29.45 | |||
| ABCB1(MDR1) | Compound 1 | 28.31 | 0.04 | 0.001 * |
| Compound 2 | 29.56 | 0.01 | ||
| Compound 3 | 27.80 | 0.08 | ||
| Control | 27.45 | |||
| GAPDH | Compound 1 | 25.27 | 1.00 | |
| Compound 2 | 23.81 | 1.00 | ||
| Compound 3 | 25.90 | 1.00 | ||
| Control | 29.14 | 1.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ozturk, A.; Agbektas, T.; Huseynzada, A.; Guliyev, R.; Ganbarova, R.; Hasanova, U.; Tas, A.; Erkan, S.; Zontul, C.; Inandiklioglu, N.; et al. In Silico and In Vitro Studies of Novel Azomethines on DNA Repair Genes in Gastric Cell Lines. Life 2023, 13, 1982. https://doi.org/10.3390/life13101982
Ozturk A, Agbektas T, Huseynzada A, Guliyev R, Ganbarova R, Hasanova U, Tas A, Erkan S, Zontul C, Inandiklioglu N, et al. In Silico and In Vitro Studies of Novel Azomethines on DNA Repair Genes in Gastric Cell Lines. Life. 2023; 13(10):1982. https://doi.org/10.3390/life13101982
Chicago/Turabian StyleOzturk, Alpaslan, Tugba Agbektas, Alakbar Huseynzada, Ruslan Guliyev, Rana Ganbarova, Ulviyya Hasanova, Ayca Tas, Sultan Erkan, Cemile Zontul, Nihal Inandiklioglu, and et al. 2023. "In Silico and In Vitro Studies of Novel Azomethines on DNA Repair Genes in Gastric Cell Lines" Life 13, no. 10: 1982. https://doi.org/10.3390/life13101982







