Study of the Anticancer Potential of Plant Extracts Using Liver Tumor Microphysiological System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fabrication of Microfluidic Device
2.2. Cell Culture and Seeding on the Microfluidic Chip
2.3. Collection and Preparation of Plant Extracts
2.4. Live/Dead Assay and ROS Estimation Assay
2.5. Biomarker Analysis
2.6. Statistical Analysis
3. Results
3.1. Real-Time Monitoring of Liver Tumor MPS
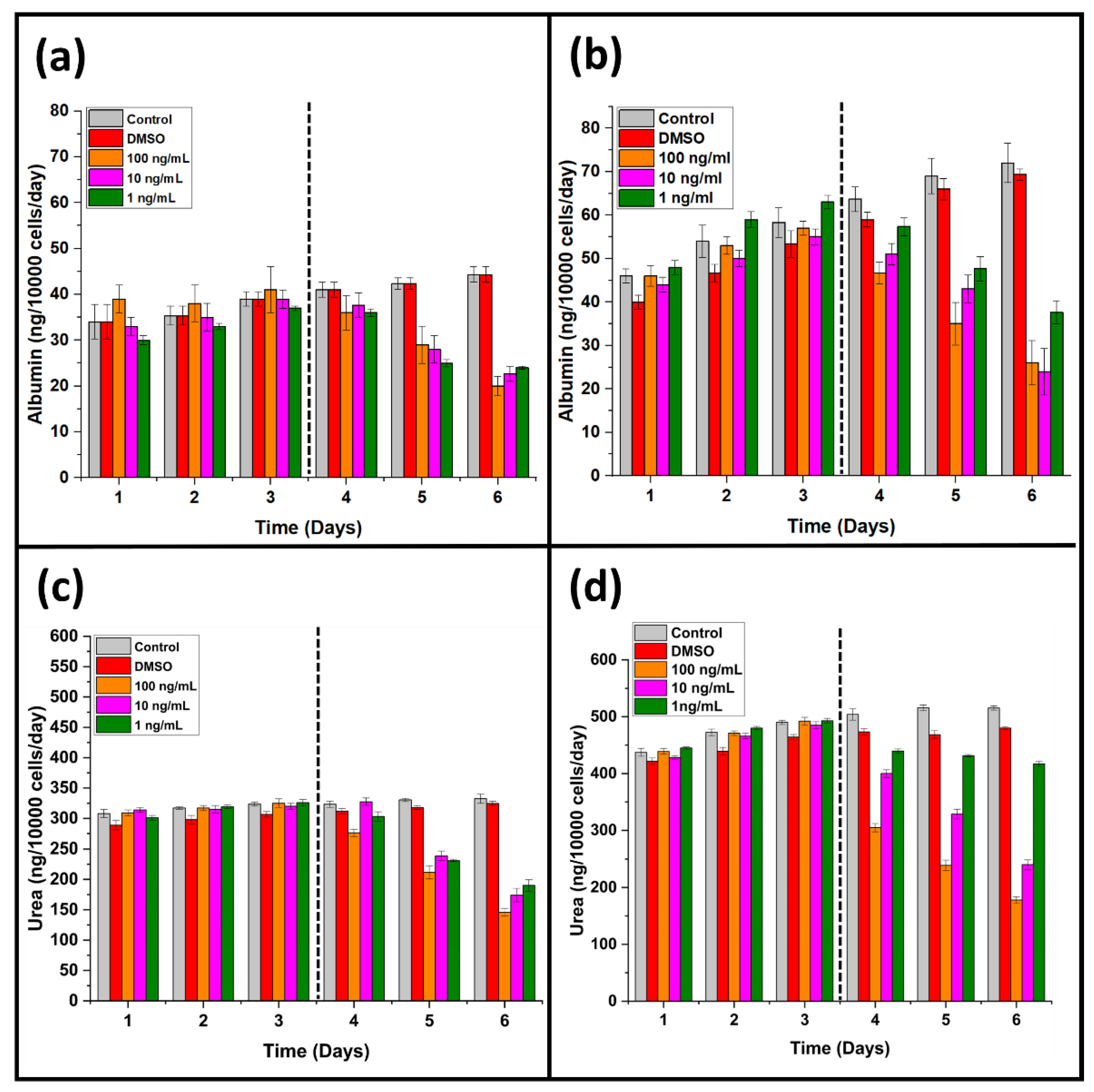
3.2. Effect of A. cappadocicum on Liver Function Tests
3.3. Comparative Analysis of Cell Viability
3.4. Effects of A. cappadocicum on Liver Tumor MPS
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef]
- Benedict, M.; Zhang, X. Non-alcoholic fatty liver disease: An expanded review. World J. Hepatol. 2017, 9, 715. [Google Scholar] [CrossRef]
- Tan, S.-L.; Pause, A.; Shi, Y.; Sonenberg, N. Hepatitis C therapeutics: Current status and emerging strategies. Nat. Rev. Drug Discov. 2002, 1, 867–881. [Google Scholar] [CrossRef]
- Yasin, S.; Bhatti, T.; Farooqi, M.U.; Mateen, F. Immunologic aspect in diagnosis and treatment of SARS-COV-2 patients. J. Shifa Tameer-E-Millat Univ. 2020, 3, 113–121. [Google Scholar] [CrossRef]
- Leontaridou, M.; Gabbert, S.; Landsiedel, R. The impact of precision uncertainty on predictive accuracy metrics of non-animal testing methods. ALTEX-Altern. Anim. Exp. 2019, 36, 435–446. [Google Scholar] [CrossRef]
- Park, T.-M.; Kang, D.; Jang, I.; Yun, W.-S.; Shim, J.-H.; Jeong, Y.H.; Kwak, J.-Y.; Yoon, S.; Jin, S. Fabrication of in vitro cancer microtissue array on fibroblast-layered nanofibrous membrane by inkjet printing. Int. J. Mol. Sci. 2017, 18, 2348. [Google Scholar] [CrossRef] [Green Version]
- Kratz, S.R.A.; Höll, G.; Schuller, P.; Ertl, P.; Rothbauer, M. Latest trends in biosensing for microphysiological organs-on-a-chip and body-on-a-chip systems. Biosensors 2019, 9, 110. [Google Scholar] [CrossRef] [Green Version]
- Salih, A.R.C.; Farooqi, H.M.U.; Kim, Y.S.; Lee, S.H.; Choi, K.H. Impact of serum concentration in cell culture media on tight junction proteins within a multiorgan microphysiological system. Microelectron. Eng. 2020, 232, 111405. [Google Scholar] [CrossRef]
- Zoio, P.; Lopes-Ventura, S.; Oliva, A. Barrier-on-a-Chip with a Modular Architecture and Integrated Sensors for Real-Time Measurement of Biological Barrier Function. Micromachines 2021, 12, 816. [Google Scholar] [CrossRef]
- Farooqi, H.M.U.; Khalid, M.A.U.; Kim, K.H.; Lee, S.R.; Choi, K.H. Real-time physiological sensor-based liver-on-chip device for monitoring drug toxicity. J. Micromechanics Microengineering 2020, 30, 115013. [Google Scholar] [CrossRef]
- Farooqi, H.M.U.; Kang, B.; Khalid, M.A.U.; Salih, A.R.C.; Hyun, K.; Park, S.H.; Huh, D.; Choi, K.H. Real-time monitoring of liver fibrosis through embedded sensors in a microphysiological system. Nano Converg. 2021, 8, 3. [Google Scholar] [CrossRef]
- Kintzios, S.E. Terrestrial plant-derived anticancer agents and plant species used in anticancer research. Crit. Rev. Plant Sci. 2006, 25, 79–113. [Google Scholar] [CrossRef]
- Kuete, V.; Eyong, K.; Folefoc, G.; Beng, V.; Hussain, H.; Krohn, K.; Nkengfack, A. Antimicrobial activity of the methanolic extract and of the chemical constituents isolated from Newbouldia laevis. Die Pharm.-Int. J. Pharm. Sci. 2007, 62, 552–556. [Google Scholar]
- Shad, A.A.; Ahmad, S.; Ullah, R.; AbdEl-Salam, N.M.; Fouad, H.; Rehman, N.U.; Hussain, H.; Saeed, W. Phytochemical and biological activities of four wild medicinal plants. Sci. World J. 2014, 2014, 857363. [Google Scholar] [CrossRef] [Green Version]
- Das, M.; Shrestha, B.; Datta, S.; Das, S.; Deb, J. Phytopharmacological review of Bacopa monnieri Linn. Nat. Prod. Anindian J. 2010, 6, 1–4. [Google Scholar]
- Ajaib, M.; Ishtiaq, M.; Bhatti, K.H.; Hussain, I.; Maqbool, M.; Hussain, T.; Mushtaq, W.; Ghani, A.; Azeem, M.; Khan, S.M.R. Inventorization of traditional ethnobotanical uses of wild plants of Dawarian and Ratti Gali areas of District Neelum, Azad Jammu and Kashmir Pakistan. PLoS ONE 2021, 16, e0255010. [Google Scholar] [CrossRef]
- Sher, Z.; Khan, Z.; Hussain, F. Ethnobotanical studies of some plants of Chagharzai valley, district Buner, Pakistan. Pak. J. Bot. 2011, 43, 1445–1452. [Google Scholar]
- Saqib, Z.; Sultan, A. Ethnobotany of Palas valley, Pakistan. Ethnobot. Leafl. 2005, 2004, 11. [Google Scholar]
- Kausar, F.; Farooqi, M.-A.; Farooqi, H.-M.-U.; Salih, A.-R.-C.; Khalil, A.-A.-K.; Kang, C.-w.; Mahmoud, M.H.; Batiha, G.-E.-S.; Choi, K.-h.; Mumtaz, A.-S. Phytochemical Investigation, Antimicrobial, Antioxidant and Anticancer Activities of Acer cappadocicum Gled. Life 2021, 11, 656. [Google Scholar] [CrossRef]
- Hazrat, A.; Wahab, M. Threatened native plants of Dir Kohistan valley, Khyber Pukhtunkhwa, Pakistan. FUUAST J. Biol. 2011, 1, 35–38. [Google Scholar]
- Kausar, F.; Kim, K.-H.; Farooqi, H.M.U.; Farooqi, M.A.; Kaleem, M.; Waqar, R.; Khalil, A.A.K.; Khuda, F.; Abdul Rahim, C.S.; Hyun, K. Evaluation of Antimicrobial and Anticancer Activities of Selected Medicinal Plants of Himalayas, Pakistan. Plants 2022, 11, 48. [Google Scholar] [CrossRef] [PubMed]
- El-Ali, J.; Sorger, P.K.; Jensen, K.F. Cells on chips. Nature 2006, 442, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Hansmann, J.; Egger, D.; Kasper, C. Advanced Dynamic Cell and Tissue Culture. Bioengineering 2018, 5, 65. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, S.T.; Nguyen, H.T.-L.; Truong, K.D. Comparative cytotoxic effects of methanol, ethanol and DMSO on human cancer cell lines. Biomed. Res. Ther. 2020, 7, 3855–3859. [Google Scholar] [CrossRef]
- Ouedraogo, V.; Kiendrebeogo, M. Methanol extract from Anogeissus leiocarpus (DC) Guill. et Perr.(Combretaceae) stem bark quenches the quorum sensing of Pseudomonas aeruginosa PAO1. Medicines 2016, 3, 26. [Google Scholar] [CrossRef] [Green Version]
- Martin, T.A.; Jiang, W.G. Loss of tight junction barrier function and its role in cancer metastasis. Biochim. Biophys. Acta (BBA)-Biomembr. 2009, 1788, 872–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, S.T.; Hwang, J.H.; Kim, D.H.; Park, M.J.; Lee, I.H.; Nam, H.J.; Kang, J.K.; Kim, S.K.; Hwang, J.S.; Chung, H.K. Role of NADH: Quinone oxidoreductase-1 in the tight junctions of colonic epithelial cells. BMB Rep. 2014, 47, 494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, F.-J.; Han, Z.; Ma, J.-H.; Jiang, S.-Q.; Zhao, X.-M.; Ruan, H.; Xie, W.-D.; Li, X. Involvement of Reactive Oxygen Species in the Hepatorenal Toxicity of Actinomycin V In Vitro and In Vivo. Mar. Drugs 2020, 18, 428. [Google Scholar] [CrossRef] [PubMed]
- Anadón, A.; Castellano, V.; Martínez-Larrañaga, M.R. Biomarkers of drug toxicity. In Biomarkers in Toxicology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 593–607. [Google Scholar]
- Deng, J.; Wei, W.; Chen, Z.; Lin, B.; Zhao, W.; Luo, Y.; Zhang, X. Engineered liver-on-a-chip platform to mimic liver functions and its biomedical applications: A review. Micromachines 2019, 10, 676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakolish, C.; Reese, C.E.; Luo, Y.-S.; Valdiviezo, A.; Schurdak, M.E.; Gough, A.; Taylor, D.L.; Chiu, W.A.; Vernetti, L.A.; Rusyn, I. Analysis of reproducibility and robustness of a human microfluidic four-cell liver acinus microphysiology system (LAMPS). Toxicology 2021, 448, 152651. [Google Scholar] [CrossRef]
- Gunasekaran, V.; Elangovan, K.; Devaraj, S.N. Targeting hepatocellular carcinoma with piperine by radical-mediated mitochondrial pathway of apoptosis: An in vitro and in vivo study. Food Chem. Toxicol. 2017, 105, 106–118. [Google Scholar] [CrossRef]
- Cai, X.; Yan, A.; Fu, N.; Wang, S. In vitro antioxidant activities of enzymatic hydrolysate from Schizochytrium Sp. and its hepatoprotective effects on acute alcohol-induced liver injury in vivo. Mar. Drugs 2017, 15, 115. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Yan, L.; Guo, L.; Sun, H.; Huang, Q.; Shao, D.; Jiang, C.; Shi, J. Effects of Bacillus subtilis iturin A on HepG2 cells in vitro and vivo. AMB Express 2021, 11, 67. [Google Scholar] [CrossRef]
- Bonnier, F.; Keating, M.; Wrobel, T.P.; Majzner, K.; Baranska, M.; Garcia-Munoz, A.; Blanco, A.; Byrne, H.J. Cell viability assessment using the Alamar blue assay: A comparison of 2D and 3D cell culture models. Toxicol. Vitr. 2015, 29, 124–131. [Google Scholar] [CrossRef] [Green Version]
- Rampersad, S.N. Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors 2012, 12, 12347–12360. [Google Scholar] [CrossRef] [PubMed]
- Single, A.; Beetham, H.; Telford, B.J.; Guilford, P.; Chen, A. A comparison of real-time and endpoint cell viability assays for improved synthetic lethal drug validation. J. Biomol. Screen. 2015, 20, 1286–1293. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, M.C.; Bolton, W.E. Comparison of cell viability probes compatible with fixation and permeabilization for combined surface and intracellular staining in flow cytometry. Cytom. J. Int. Soc. Anal. Cytol. 1995, 19, 243–255. [Google Scholar] [CrossRef]
- Huh, D.; Torisawa, Y.-s.; Hamilton, G.A.; Kim, H.J.; Ingber, D.E. Microengineered physiological biomimicry: Organs-on-chips. Lab Chip 2012, 12, 2156–2164. [Google Scholar] [CrossRef]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Tsao, C.-W.; Cheng, Y.-C.; Cheng, J.-H. Fluid flow shear stress stimulation on a multiplex microfluidic device for rat bone marrow stromal cell differentiation enhancement. Micromachines 2015, 6, 1996–2009. [Google Scholar] [CrossRef]
- Chethikkattuveli Salih, A.R.; Hyun, K.; Asif, A.; Soomro, A.M.; Farooqi, H.M.U.; Kim, Y.S.; Kim, K.H.; Lee, J.W.; Huh, D.; Choi, K.H. Extracellular Matrix Optimization for Enhanced Physiological Relevance in Hepatic Tissue-Chips. Polymers 2021, 13, 3016. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.J.; Guntur, S.R.; Lee, K.I.; Paeng, D.G.; Coleman, A. An optically transparent tissue mimicking phantom for monitoring the thermal lesion produced by high intensity focused ultrasound. J. Acoust. Soc. Am. 2012, 131, 3364. [Google Scholar] [CrossRef]
- Yeom, E.; Nam, K.-H.; Paeng, D.-G.; Lee, S.-J. Effects of red blood cell aggregates dissociation on the estimation of ultrasound speckle image velocimetry. Ultrasonics 2014, 54, 1480–1487. [Google Scholar] [CrossRef]
- Moloney, J.N.; Cotter, T.G. ROS signalling in the biology of cancer. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Milkovic, L.; Cipak Gasparovic, A.; Cindric, M.; Mouthuy, P.-A.; Zarkovic, N. Short overview of ROS as cell function regulators and their implications in therapy concepts. Cells 2019, 8, 793. [Google Scholar] [CrossRef] [Green Version]
- Griess, B.; Tom, E.; Domann, F.; Teoh-Fitzgerald, M. Extracellular superoxide dismutase and its role in cancer. Free Radic. Biol. Med. 2017, 112, 464–479. [Google Scholar] [CrossRef] [PubMed]
- Case, A.J. On the origin of superoxide dismutase: An evolutionary perspective of superoxide-mediated redox signaling. Antioxidants 2017, 6, 82. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farooqi, H.M.U.; Sammantasinghar, A.; Kausar, F.; Farooqi, M.A.; Chethikkattuveli Salih, A.R.; Hyun, K.; Lim, J.-H.; Khalil, A.A.K.; Mumtaz, A.S.; Choi, K.H. Study of the Anticancer Potential of Plant Extracts Using Liver Tumor Microphysiological System. Life 2022, 12, 135. https://doi.org/10.3390/life12020135
Farooqi HMU, Sammantasinghar A, Kausar F, Farooqi MA, Chethikkattuveli Salih AR, Hyun K, Lim J-H, Khalil AAK, Mumtaz AS, Choi KH. Study of the Anticancer Potential of Plant Extracts Using Liver Tumor Microphysiological System. Life. 2022; 12(2):135. https://doi.org/10.3390/life12020135
Chicago/Turabian StyleFarooqi, Hafiz Muhammad Umer, Anupamma Sammantasinghar, Farzana Kausar, Muhammad Awais Farooqi, Abdul Rahim Chethikkattuveli Salih, Kinam Hyun, Jong-Hwan Lim, Atif Ali Khan Khalil, Abdul Samad Mumtaz, and Kyung Hyun Choi. 2022. "Study of the Anticancer Potential of Plant Extracts Using Liver Tumor Microphysiological System" Life 12, no. 2: 135. https://doi.org/10.3390/life12020135






