The Performance and Mechanism of Sludge Reduction by the Bioaugmentation Approach
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Enriching
2.2. Isolation and Identification
2.3. Sludge Reduction Experiments
2.4. Physicochemical Analysis
3. Results and Discussion
3.1. Isolation and Identification of Sludge-Reductants
| Name | Accession No. | Top-Hit Strain | Top-Hit Phylum | Similarity (%) | Completeness (%) |
|---|---|---|---|---|---|
| WAS-3 a-5-1 | OP379642 | Humibacter ginsengiterrae DCY60(T) | Actinobacteria | 93.1 | 98.1 |
| WAS-3 a-5-2 | OP379643 | Xinfangfangia soli ZQBW(T) | Proteobacteria | 97.4 | 100 |
| WAS-3 a-10-1 | OP379637 | Flexivirga lutea TBS-100(T) | Actinobacteria | 91.8 | 95.6 |
| WAS-3 a-10-2 | OP379638 | Novosphingobium arabidopsis CC-ALB-2(T) | Proteobacteria | 94.4 | 100 |
| WAS-3 a-10-3 | OP379639 | Microbacterium flavum YM18-098(T) | Actinobacteria | 94.9 | 99.0 |
| WAS-3 a-11-1 | OP379640 | Kocuria salina Hv14b(T) | Actinobacteria | 95.2 | 96.8 |
| WAS-3 a-12-1 | OP379641 | Noviherbaspirillum soli SUEMI10(T) | Proteobacteria | 90.7 | 100 |
| WAS-7 a-8-1 | OP379646 | Rhodanobacter terrae GP18-1(T) | Proteobacteria | 98.6 | 100 |
| WAS-7 a-10-1 | OP379644 | Staphylococcus warneri ATCC 27836(T) | Firmicutes | 91.9 | 99.7 |
| WAS-7 a-10-3 | OP379645 | Cellulomonas massiliensis JC225(T) | Actinobacteria | 92.3 | 100 |
| WAS-10 a-6-1 | OP379632 | Diaphorobacter oryzae RF3(T) | Proteobacteria | 95.1 | 96.6 |
| WAS-10 a-6-2 | OP379633 | Bacillus aryabhattai B8W22(T) | Firmicutes | 94.0 | 100 |
| WAS-10 a-9-1 | OP379634 | Bacillus sporothermodurans M215(T) | Firmicutes | 88.9 | 99.5 |
| WAS-10 a-9-2 | OP379635 | Bacillus vini LAM0415(T) | Firmicutes | 88.7 | 99.1 |
| WAS-10 a-9-3 | OP379636 | Bacillus cereus ATCC 14579(T) | Firmicutes | 99.2 | 100 |
| WAS-10 a-10-1 | OP379629 | Luteimonas cucumeris CGMCC 1.10821(T) | Proteobacteria | 93.1 | 100 |
| WAS-10 a-12-1 | OP379630 | Bacillus oryzaecorticis R1(T) | Firmicutes | 96.3 | 75.6 |
| WAS-10 a-12-2 | OP379631 | Ottowia pentelensis RB3-7(T) | Proteobacteria | 92.1 | 100 |
| WAS-UT b-2 | OP379647 | Bacillus anthracis Ames (T) | Firmicutes | 99.2 | 100 |
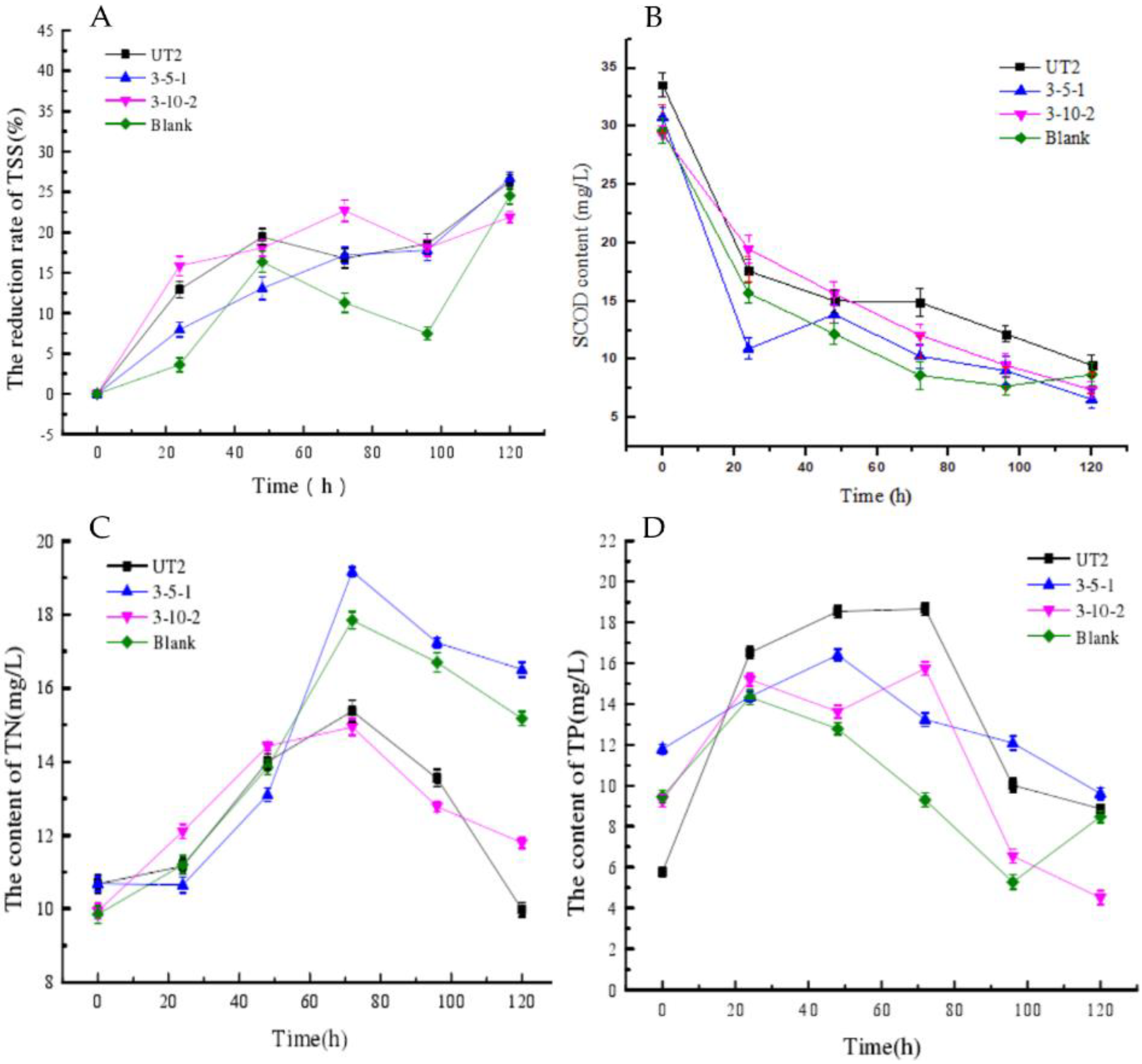
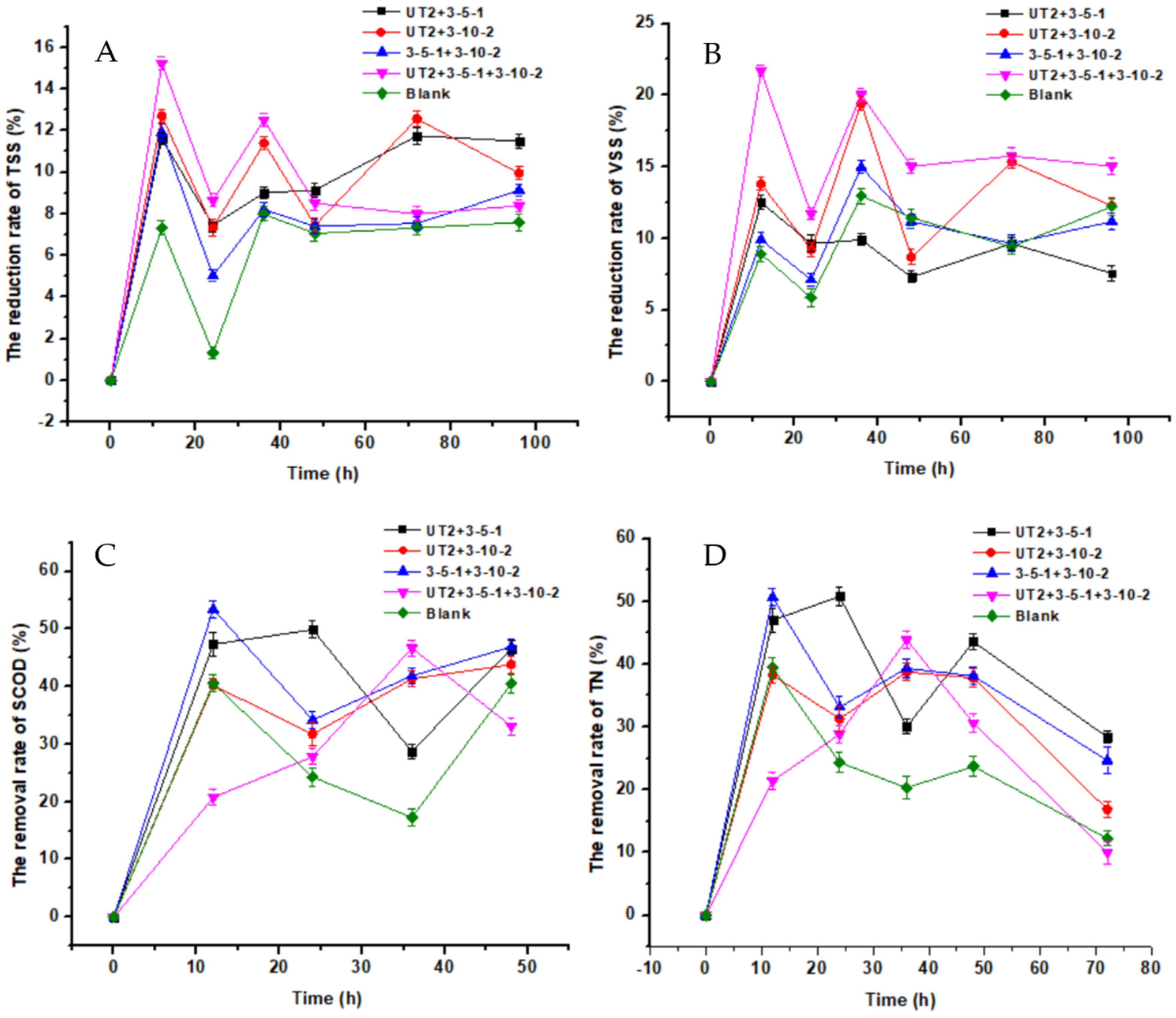
3.2. Sludge Reduction by Pure or Mixed Cultures
3.3. Optimization of Sludge Reduction
3.4. The Study of the Sludge Reduction Mechanism
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Machineni, L. Review on biological wastewater treatment and resources recovery: Attached and suspended growth systems. Water Sci. Technol. 2019, 80, 2013–2026. [Google Scholar] [CrossRef]
- Zhu, G.; Peng, Y.; Li, B.; Guo, J.; Yang, Q.; Wang, S. Biological removal of nitrogen from wastewater. Rev. Environ. Contam. Toxicol. 2008, 192, 159–195. [Google Scholar]
- Zhang, Q.; Yang, W.; Ngo, H.; Guo, W.; Jin, P.; Dzakpasu, M.; Yang, S.; Wang, Q.; Wang, X.; Ao, D. Current status of urban wastewater treatment plants in China. Environ. Int. 2016, 92, 11–22. [Google Scholar] [CrossRef]
- Petroody, S.S.A.; Hashemi, S.H.; van Gestel, C.A. Transport and accumulation of microplastics through wastewater treatment sludge processes. Chemosphere 2021, 278, 130471. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Chen, Q.; van Loosdrecht, M.C.; Li, J.; Jiang, H. Sustainable disposal of excess sludge: Incineration without anaerobic digestion. Water Res. 2020, 170, 115298. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.-A.; Mika, S.; Pirjo, I. Sewage sludge electro-dewatering treatment—A review. Dry. Technol. 2012, 30, 691–706. [Google Scholar] [CrossRef]
- Skinner, S.J.; Studer, L.J.; Dixon, D.R.; Hillis, P.; Rees, C.A.; Wall, R.C.; Cavalida, R.G.; Usher, S.P.; Stickland, A.D.; Scales, P.J. Quantification of wastewater sludge dewatering. Water Res. 2015, 82, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Xiao, K.; Zhu, W.; Withanage, N.; Zhou, Y. Enhanced sludge solubilization and dewaterability by synergistic effects of nitrite and freezing. Water Res. 2018, 130, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Appels, L.; Su, H. Combining microwave irradiation with sodium citrate addition improves the pre-treatment on anaerobic digestion of excess sewage sludge. J. Environ. Manag. 2018, 213, 271–278. [Google Scholar] [CrossRef]
- Donoso-Bravo, A.; Pérez-Elvira, S.; Fdz-Polanco, F. Application of simplified models for anaerobic biodegradability tests. Evaluation of pre-treatment processes. Chem. Eng. J. 2010, 160, 607–614. [Google Scholar] [CrossRef]
- Wang, Q.; Wei, W.; Gong, Y.; Yu, Q.; Li, Q.; Sun, J.; Yuan, Z. Technologies for reducing sludge production in wastewater treatment plants: State of the art. Sci. Total Environ. 2017, 587, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, D.; Zhang, G.; Ren, Z.; Zhu, Y. Critical review on ultrasound lysis-cryptic growth for sludge reduction. J. Environ. Chem. Eng. 2021, 9, 106263. [Google Scholar] [CrossRef]
- Lan, W.; Li, Y.; Bi, Q.; Hu, Y. Reduction of excess sludge production in sequencing batch reactor (SBR) by lysis–cryptic growth using homogenization disruption. Bioresour. Technol. 2013, 134, 43–50. [Google Scholar] [CrossRef]
- Ma, H.; Zhang, S.; Lu, X.; Xi, B.; Guo, X.; Wang, H.; Duan, J. Excess sludge reduction using pilot-scale lysis-cryptic growth system integrated ultrasonic/alkaline disintegration and hydrolysis/acidogenesis pretreatment. Bioresour. Technol. 2012, 116, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Toya, S.; Sabidi, S.; Hoshiko, Y.; Maeda, T. Diluted Luria-Bertani medium vs. sewage sludge as growth media: Comparison of community structure and diversity in the culturable bacteria. Appl. Microbiol. Biotechnol. 2021, 105, 3787–3798. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Guo, J.; Li, L.; Asem, M.D.; Zhang, Y.; Mohamad, O.A.; Salam, N.; Li, W. Endophytic bacteria associated with endangered plant Ferula sinkiangensis K.M. Shen in an arid land: Diversity and plant growth-promoting traits. J. Arid. Land 2017, 9, 432–445. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Zhang, Z.; Tsapekos, P.; Alvarado-Morales, M.; Zhu, X.; Zervas, A.; Jacobsen, C.S.; Angelidaki, I. Enhanced fermentative lactic acid production from source-sorted organic household waste: Focusing on low-pH microbial adaptation and bio-augmentation strategy. Sci. Total Environ. 2022, 808, 152129. [Google Scholar] [CrossRef]
- Foladori, P.; Bruni, L.; Tamburini, S.; Ziglio, G. Direct quantification of bacterial biomass in influent, effluent and activated sludge of wastewater treatment plants by using flow cytometry. Water Res. 2010, 44, 3807–3818. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhang, G.; Li, J.; Zhao, Z.; Kang, X. Effect of endogenous hydrolytic enzymes pretreatment on the anaerobic digestion of sludge. Bioresour. Technol. 2013, 146, 758–761. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Guo, L.; Li, Q.; Zhao, Y.; Gao, M.; She, Z.; Jin, C. Three-dimensional fluorescence excitation–emission matrix (EEM) spectroscopy with regional integration analysis for assessing waste sludge hydrolysis at different pretreated temperatures. Environ. Sci. Pollut. Res. 2016, 23, 24061–24067. [Google Scholar] [CrossRef]
- Guo, J.; Ni, B.-J.; Han, X.; Chen, X.; Bond, P.; Peng, Y.; Yuan, Z. Unraveling microbial structure and diversity of activated sludge in a full-scale simultaneous nitrogen and phosphorus removal plant using metagenomic sequencing. Enzym. Microb. Technol. 2017, 102, 16–25. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, W.; Liu, R.; Li, Q.; Li, B.; Wang, S.; Song, C.; Qiao, C.; Mulchandani, A. Phylogenetic diversity and metabolic potential of activated sludge microbial communities in full-scale wastewater treatment plants. Environ. Sci. Technol. 2011, 45, 7408–7415. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ruhyadi, R.; Huang, J.; Yan, W.; Wang, G.; Shen, N.; Hanggoro, W. Comprehensive comparison of acidic and alkaline anaerobic fermentations of waste activated sludge. Bioresour. Technol. 2021, 323, 124613. [Google Scholar] [CrossRef] [PubMed]
- Bian, B.; Zhang, L.; Zhang, Q.; Zhang, S.; Yang, Z.; Yang, W. Coupled heating/acidification pretreatment of chemical sludge for dewatering by using waste sulfuric acid at low temperature. Chemosphere 2018, 205, 260–266. [Google Scholar] [CrossRef]
- Dhall, P.; Kumar, R.; Kumar, A. Biodegradation of sewage wastewater using Autochthonous bacteria. Sci. World J. 2012, 2012, 861903. [Google Scholar] [CrossRef] [Green Version]
- Seo, K.W.; Choi, Y.-S.; Gu, M.B.; Kwon, E.E.; Tsang, Y.F.; Rinklebe, J.; Park, C. Pilot-scale investigation of sludge reduction in aerobic digestion system with endospore-forming bacteria. Chemosphere 2017, 186, 202–208. [Google Scholar] [CrossRef]
- Gao, Y.; Peng, Y.; Zhang, J.; Wang, S.; Guo, J.; Ye, L. Biological sludge reduction and enhanced nutrient removal in a pilot-scale system with 2-step sludge alkaline fermentation and A2O process. Bioresour. Technol. 2011, 102, 4091–4097. [Google Scholar] [CrossRef]
- Moser-Engeler, R.; Udert, K.M.; Wild, D.; Siegrist, H. Products from primary sludge fermentation and their suitability for nutrient removal. Water Sci.Technol. 1998, 38, 265–273. [Google Scholar] [CrossRef]
- Liébana, R.; Arregui, L.; Santos, A.; Murciano, A.; Marquina, D.; Serrano, S. Unravelling the interactions among microbial populations found in activated sludge during biofilm formation. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [PubMed]
- Rocher, M.; Roux, G.; Goma, G.; Pilas Begue, A.; Louvel, L.; Rols, J. Excess sludge reduction in activated sludge processes by integrating biomass alkaline heat treatment. Water Sci. Technol. 2011, 44, 437–444. [Google Scholar] [CrossRef]
- Uma Rani, R.; Adish Kumar, S.; Kaliappan, S.; Yeom, I.-T.; Rajesh Banu, J. Low temperature thermo-chemical pretreatment of dairy waste activated sludge for anaerobic digestion process. Bioresour. Technol. 2012, 103, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Wang, X.; Xu, H.; He, P. Enhanced anaerobic digestion and sludge dewaterability by alkaline pretreatment and its mechanism. J. Environ. Sci. 2012, 24, 1731–1738. [Google Scholar] [CrossRef]
- Jiang, L.-M.; Zhou, Z.; Cheng, C.; Li, J.; Huang, C.; Niu, T. Sludge reduction by a micro-aerobic hydrolysis process: A full-scale application and sludge reduction mechanisms. Bioresour. Technol. 2018, 268, 684–691. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Yoshimura, T.; García-Contreras, R.; Ogawa, H.I. Purification and characterization of a serine protease secreted by Brevibacillus sp. KH3 for reducing waste activated sludge and biofilm formation. Bioresour. Technol. 2011, 102, 10650–10656. [Google Scholar] [CrossRef]
- Yang, Q.; Luo, K.; Li, X.M.; Wang, D.B.; Zheng, W.; Zeng, G.M.; Liu, J.J. Enhanced efficiency of biological excess sludge hydrolysis under anaerobic digestion by additional enzymes. Bioresour. Technol. 2010, 101, 2924–2930. [Google Scholar] [CrossRef]
- Lin, F.; Zhu, X.; Li, J.; Yu, P.; Luo, Y.; Liu, M. Effect of extracellular polymeric substances (EPS) conditioned by combined lysozyme and cationic polyacrylamide on the dewatering performance of activated sludge. Chemosphere 2019, 235, 679–689. [Google Scholar] [CrossRef]

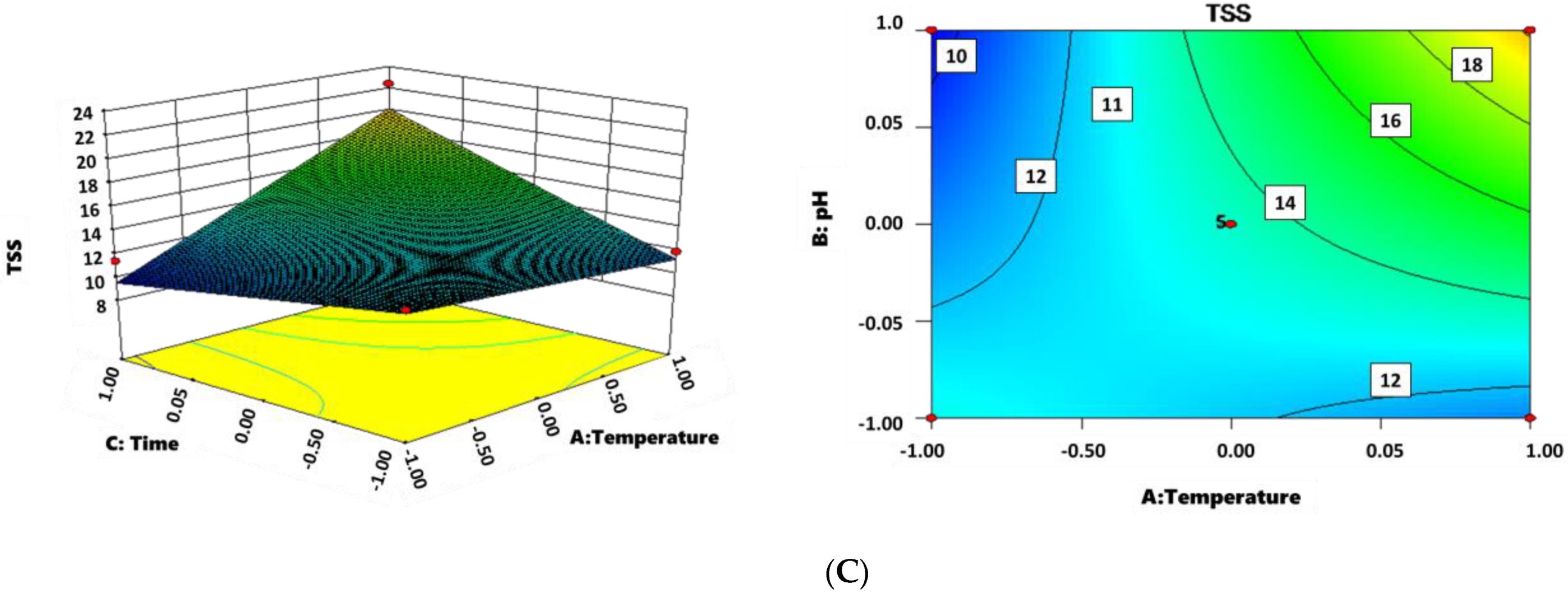


| No. | X1 a (Temperature) | X2 b (pH) | X3 c (Incubation Time) | TSS Removal (%) |
|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 13.44 |
| 2 | −1 | 0 | −1 | 22.41 |
| 3 | −1 | 0 | 1 | 11.87 |
| 4 | 0 | −1 | −1 | 13.51 |
| 5 | 1 | 0 | −1 | 11.40 |
| 6 | 0 | −1 | 1 | 10.60 |
| 7 | 0 | 0 | 0 | 13.44 |
| 8 | 0 | 1 | −1 | 14.24 |
| 9 | 1 | 0 | 1 | 13.24 |
| 10 | 1 | −1 | 0 | 9.53 |
| 11 | −1 | 1 | 0 | 13.71 |
| 12 | 0 | 0 | 0 | 13.44 |
| 13 | 1 | 1 | 0 | 11.27 |
| 14 | 0 | 0 | 0 | 11.27 |
| 15 | 0 | 0 | 0 | 11.27 |
| 16 | −1 | −1 | 0 | 15.21 |
| 17 | 0 | 1 | 1 | 14.98 |
| Source of Variation | Sum of Squares | Degree | of Mean | F-Value | Prob > F | Significance |
|---|---|---|---|---|---|---|
| Equation | 102.01 | 6 | 17.00 | 8.80 | 0.0016 | significant |
| X1 | 39.42 | 1 | 39.42 | 20.4 | 0.0011 | — |
| X2 | 3.58 | 1 | 3.58 | 1.85 | 0.2034 | — |
| X3 | 14.77 | 1 | 14.77 | 7.64 | 0.0200 | — |
| X1X2 | 2.63 | 1 | 2.63 | 1.36 | 0.2704 | — |
| X1X3 | 38.28 | 1 | 38.28 | 19.81 | 0.0012 | — |
| X2X3 | 3.32 | 1 | 3.32 | 1.72 | 0.2191 | — |
| Residual | 19.33 | 10 | 1.93 | — | — | — |
| Lack of fit analysis | 19.33 | 6 | 3.22 | — | 0.1819 | not significant |
| Pure error | 0 | 4 | 0.000 | — | — | — |
| Sum | 121.33 | 16 | — | — | — | — |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Yang, X.; Hu, A.; Li, Y.; Li, Y.; Fu, L.; Yu, C.-P. The Performance and Mechanism of Sludge Reduction by the Bioaugmentation Approach. Life 2022, 12, 1649. https://doi.org/10.3390/life12101649
Li J, Yang X, Hu A, Li Y, Li Y, Fu L, Yu C-P. The Performance and Mechanism of Sludge Reduction by the Bioaugmentation Approach. Life. 2022; 12(10):1649. https://doi.org/10.3390/life12101649
Chicago/Turabian StyleLi, Jiangwei, Xiaoyong Yang, Anyi Hu, Yan Li, Yeyun Li, Lijun Fu, and Chang-Ping Yu. 2022. "The Performance and Mechanism of Sludge Reduction by the Bioaugmentation Approach" Life 12, no. 10: 1649. https://doi.org/10.3390/life12101649






